Weakly Supervised Contrastive Learning for Chest X-Ray Report Generation
Abstract
Radiology report generation aims at generating descriptive text from radiology images automatically, which may present an opportunity to improve radiology reporting and interpretation. A typical setting consists of training encoder-decoder models on image-report pairs with a cross entropy loss, which struggles to generate informative sentences for clinical diagnoses since normal findings dominate the datasets. To tackle this challenge and encourage more clinically-accurate text outputs, we propose a novel weakly supervised contrastive loss for medical report generation. Experimental results demonstrate that our method benefits from contrasting target reports with incorrect but semantically-close ones. It outperforms previous work on both clinical correctness and text generation metrics for two public benchmarks.
1 Introduction
Automated radiology report generation aims at generating informative text from radiologic image studies. It could potentially improve radiology reporting and alleviate the workload of radiologists. Recently, following the success of deep learning in conditional text generation tasks such as image captioning (Vinyals et al., 2015; Xu et al., 2015), many methods have been proposed for this task (Jing et al., 2017; Li et al., 2018; Liu et al., 2019; Jing et al., 2020; Ni et al., 2020; Chen et al., 2020b).
Unlike conventional image captioning benchmarks (e.g. MS-COCO (Lin et al., 2014)) where referenced captions are usually short, radiology reports are much longer with multiple sentences, which pose higher requirements for information selection, relation extraction, and content ordering. To generate informative text from a radiology image study, a caption model is required to understand the content, identify abnormal positions in an image and organize the wording to describe findings in images. However, the standard approach of training an encoder-decoder model with teacher forcing and cross-entropy loss often leads to text generation outputs with high frequency tokens or sentences appearing too often (Ranzato et al., 2015; Holtzman et al., 2019). This problem could be worse for chest X-ray report generation, since the task has a relatively narrow text distribution with domain-specific terminology and descriptions for medical images, and models often struggle to generate long and diverse reports (Harzig et al., 2019; Boag et al., 2020).
To tackle these challenging issues, we propose to introduce contrastive learning into chest X-ray report generation. However, simply using random non-target sequences as negative examples in a contrastive framework is suboptimal (Lee et al., 2020), as random samples are usually easy to distinguish from the correct ones. Hence, we further introduce a weakly supervised contrastive loss that assigns more weights to reports that are semantically close to the target. By exposing the model to these “hard” negative examples during training, it could learn robust representations which capture the essence of a medical image and generate high-quality reports with improved performance on clinical correctness for unseen images.
Overall, our contributions are three-fold:
-
•
We propose a novel objective for training a chest X-ray report generation model with a contrastive term, which contrasts target reports with incorrect ones during training.
-
•
We develop a weakly supervised method to identify “hard” negative samples and assign them with higher weights in our contrastive loss to further encourage diversity.
-
•
We conduct comprehensive experiments to show the effectiveness of our method, which outperforms existing methods on both clinical correctness and text generation metrics.
2 Related Work
Medical Report Generation
Medical report generation, which aims to automatically generate descriptions for clinical radiographs (e.g. chest X-rays), has drawn attention in both the machine learning and medical communities. Many methods have been proposed to solve this task. For example, Jing et al. (2017) proposed a co-attention hierarchical CNN-RNN model. Li et al. (2018, 2019) used hybrid retrieval-generation models to complement generation. Most recently, Ni et al. (2020) proposed to learn visual semantic embeddings for cross-modal retrieval in a contrastive setting, and showed improvements in identifying abnormal findings. However, this is hard to scale or generalize since we need to build template abnormal sentences for new datasets. Chen et al. (2020b) leveraged a memory-augmented transformer model to improve the ability to generate long and coherent text, but didn’t address the issue of generating dominant normal findings specifically. Our work proposes to incorporate contrastive learning into training a generation-based model, which benefits from the contrastive loss to encourage diversity and is also easy to scale compared to retrieval-based methods.
Contrastive Learning
Contrastive learning (Gutmann and Hyvärinen, 2010; Oord et al., 2018) has been widely used in many fields of machine learning. The goal is to learn a representation by contrasting positive and negative pairs. Recent work showed that contrastive learning can boost the performance of self-supervised and semi-supervised learning in computer vision tasks (He et al., 2020; Chen et al., 2020a; Khosla et al., 2020). In natural language processing, contrastive learning has been investigated for several tasks, including language modeling (Huang et al., 2018), unsupervised word alignment (Liu and Sun, 2015) and machine translation (Yang et al., 2019; Lee et al., 2020). In this work, we are interested in applying contrastive learning to chest X-ray report generation in a multi-modality setting. Different from previous work in applying contrastive learning to image captioning (Dai and Lin, 2017), we leverage a recent contrastive formulation inspired by (Chen et al., 2020a) which transforms representations into latent spaces, and propose a novel learning objective for the medical report generation task specifically.
3 Method
Generating Reports with Transformer
We leverage a memory-driven transformer proposed in (Chen et al., 2020b) as our backbone model, which uses a memory module to record key information when generating long texts.
Given a chest X-ray image , its visual features are extracted by pre-trained convolutional neural networks (e.g. ResNet (He et al., 2016)). Then we use the standard encoder in transformer to obtain hidden visual features . The decoding process at each time step can be formalized as
| (1) |
We use a cross-entropy (CE) loss to maximize the conditional log likelihood for a given observations as follows:
| (2) |
Labeling Reports with Finetuned BERT
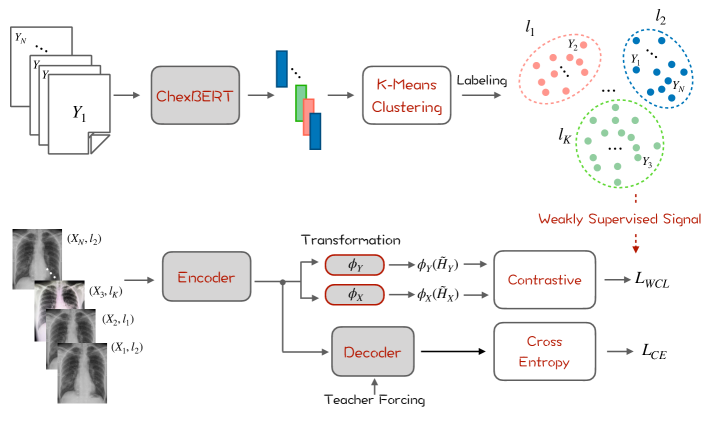
As shown in Figure 1, we first extract the embeddings of each report from ChexBERT (Smit et al., 2020), a BERT model pretrained with biomedical text and finetuned for chest X-ray report labeling. We use the [CLS] embedding of BERT to represent report-level features. We then apply K-Means to cluster the reports into groups. After clustering, each report is assigned with a corresponding cluster label , where reports in the same cluster are considered to be semantically close to each other.
Weakly supervised Contrastive Learning
To regularize the training process, we propose a weakly supervised contrastive loss (WCL). We first project the hidden representations of the image and the target sequence into a latent space:
| (3) |
where and are the average pooling of the hidden states and from the transformer, and are two fully connected layers with ReLU activation (Nair and Hinton, 2010). We then maximize the similarity between the pair of source image and target sequence, while minimizing the similarity between the negative pairs as follows:
| (4) |
where , is the cosine similarity between two vectors, is the temperature parameter, is a hyperparameter that weighs the importance of negative samples that are semantically close to the target sequence, i.e., with the same cluster label in Eq. 4. Empirically, we find that these samples are “hard” negative samples and the model would perform better by assigning more weights to distinguish these samples.
Overall, the model is optimized with a mixture of cross-entropy loss and weakly supervised contrastive loss:
| (5) |
where is a hyperparameter that weighs the two losses.
4 Experiments
| Dataset | Model | NLG metrics | CE metrics | |||||
|---|---|---|---|---|---|---|---|---|
| BLEU-1 | BLEU-4 | METEOR | ROUGE-L | Precision | Recall | F-1 | ||
| MIMIC-ABN | ST | 14.9 | 3.3 | 7.2 | 17.4 | 20.3 | 22.2 | 21.2 |
| HCR | 8.4 | 1.9 | 5.9 | 14.9 | 26.1 | 15.7 | 19.6 | |
| CVSE | 19.2 | 3.6 | 7.7 | 15.3 | 31.7 | 22.4 | 26.2 | |
| MDT | 24.6 | 6.6 | 9.7 | 23.0 | 34.0 | 29.1 | 29.4 | |
| MDT+WCL | 25.6 | 6.7 | 10.0 | 24.1 | 33.2 | 30.9 | 30.0 | |
| MIMIC-CXR | ST | 29.9 | 8.4 | 12.4 | 26.3 | 24.9 | 20.3 | 20.4 |
| TopDown | 31.7 | 9.2 | 12.8 | 26.7 | 32.0 | 23.1 | 23.8 | |
| MDT | 35.3 | 10.3 | 14.2 | 27.7 | 33.3 | 27.3 | 27.6 | |
| MDT+WCL | 37.3 | 10.7 | 14.4 | 27.4 | 38.5 | 27.4 | 29.4 | |
4.1 Experimental Setup
Datasets
We conduct experiments on two datasets: (1) MIMIC-ABN, which was proposed in (Ni et al., 2020) and contains a subset of images of MIMIC-CXR with abnormal sentences only, with 26,946/3,801/7,804 reports for train/val/test sets. (2) MIMIC-CXR (Johnson et al., 2019), the largest radiology dataset to date that consists of 222,758/1,808/3,269 reports for train/val/test sets.
Evaluation Metrics
Performance is first evaluated on three automatic metrics: BLEU (Papineni et al., 2002), ROUGE-L (Lin, 2004), and METEOR (Denkowski and Lavie, 2011).
We then use the CheXpert labeler to evaluate the clinical accuracy of the abnormal findings reported by each model, which is a state-of-the-art rule-based chest X-ray report labeling system (Irvin et al., 2019). Given sentences of abnormal findings, CheXpert will give a positive and negative label for 14 diseases. We then calculate the Precision, Recall and Accuracy for each disease based on the labels obtained from each model’s output and from the ground-truth reports.
Implementation Details
We use Adam as the optimizer with initial learning rates of 5e-5 for the visual extractor and 1e-4 for other parameters. The learning rate is decayed by a factor of 0.8 per epoch. We set the number of labels to 13 and 14 for MIMIC-ABN and MIMIC-CXR, the weighting parameters and to 0.2 and 2 respectively. We conduct a grid-based hyperparameter search for weighting factor and temperature by evaluating the models on the validation sets of the two datasets.
Words with frequency less than 3 and 10 are discarded for MIMIC-ABN and MIMIC-CXR respectively. The maximum sequence lengths are set to 64 and 100 for MIMIC-ABN and MIMIC-CXR. The projection heads consists of two convolutional layers with ReLU activation, and the latent dimensions are set to 256 for both visual and text projection layers. The hidden sizes of both the encoder and decoder are 512 with 8 heads and 3 layers. The batch size for training the two datasets is 128. We report the results using models with the highest BLEU-4 on the validation set. We use a beam size of 3 for generation to balance the generation effectiveness and efficiency.
4.2 Performance comparison
We compare our approach to other methods on two datasets. As shown in Table 1, first, our method (MDT+WCL) outperforms previous retrieval (CVSE) and generation based models (MDT) on most text generation metrics. Second, our contrastive loss significantly improves clinical efficacy metrics, demonstrating its capability to accurately report abnormal findings. Finally, the relative difference between MDT and MDT+WCL is higher on MIMIC-CXR, which contains a larger training set for learning robust representations.
4.3 Analysis
Different Contrastive Losses
| Method | Validation | |||
|---|---|---|---|---|
| BLEU-1 | BLEU-4 | METEOR | ROUGE-L | |
| baseline | 25.0 | 6.6 | 9.7 | 23.3 |
| adversarial | 25.1 | 6.6 | 9.8 | 23.2 |
| excluding | 24.8 | 6.6 | 9.8 | 23.1 |
| Ours | 25.4 | 6.8 | 10.0 | 24.0 |
We compare our contrastive formulation with other variants in Table 2. Our approach achieves the best performance over all baselines on different metrics. Both adversarial and excluding have similar performance compared to the vanilla contrastive framework (Chen et al., 2020a), not showing improvements for medical report generation. On the other hand, we identify the reports in the same cluster as “hard” negative samples and assign more weights on them during training, guiding the model to better distinguish reference reports from inaccurate ones.
Length Distributions
To further evaluate the generation quality of our method in addition to NLG and CE metrics, we compare the length of generated reports to the ground truth. To do this, we categorize all reports generated on the MIMIC-CXR test set into 20 groups (within the range of [0, 100] with an interval of 5). As shown in Figure 2, the baseline model MDT has a sharper distribution, while adding our weakly supervised contrastive loss leads to a length distribution which is smoother and closer to the ground truth, demonstrating its effectiveness to generate more diverse and accurate reports, and generalize on unseen images.

Case Study
| GT: streaky left basilar opacity likely reflects atelectasis. minimal left basilar atelectasis. history. with history of myocardial infarction presenting with epigastric pain. |
| WCL: linear left basilar opacity is likely atelectasis versus scarring. m with chest pain. |
| GT: the lungs are hyperexpanded but clear consolidation effusion or pneumothorax. increased lucency at the left lung apex and linear markings on the lateral raises the possibility apical bullous disease. hyperexpansion without acute cardiopulmonary process. |
| WCL: the lungs are hyperinflated but clear of consolidation or effusion. hyperinflation without acute cardiopulmonary process. |
| GT: there is bibasilar atelectasis. a linq cardiac monitoring device projects over the subcutaneous tissue of the left lower chest. f with shortness of breath. evaluate for pneumonia. |
| WCL: the lungs are hyperinflated with flattening of the diaphragms suggesting chronic obstructive pulmonary disease. there is mild bibasilar atelectasis. |
| GT: there is moderate amount of right-sided subcutaneous emphysema which is similar in appearance compared to prior. right-sided chest tube is again visualized. there is no increase in the pneumothorax. bilateral parenchymal opacities are again visualized and not significantly changed. the tracheostomy tube is in standard location. right subclavian line tip is in the mid svc. |
| WCL: tracheostomy tube tip is in unchanged position. right-sided port-a-cath tip terminates in the low svc. left-sided port-a-cath tip terminates in the proximal right atrium unchanged. heart size is normal. mediastinal and hilar contours are similar. innumerable bilateral pulmonary nodules are re- demonstrated better assessed on the previous ct. small right pleural effusion appears slightly increased compared to the prior exam. small left pleural effusion is similar. no new focal consolidation or pneumothorax is present. there are no acute osseous abnormalities. |
| GT: the lungs are mildly hyperinflated as evidenced by flattening of the diaphragms on the lateral view. diffuse interstitial markings compatible with known chronic interstitial lung disease are unchanged. there is no pleural effusion or evidence of pulmonary edema. there is no focal airspace consolidation worrisome for pneumonia. mild to moderate cardiomegaly is unchanged. the mediastinal and hilar contours are unremarkable. a coronary artery stent is noted. there is. levoscoliosis of the thoracic spine . |
| WCL: lung volumes are low. heart size is mildly enlarged. the aorta is tortuous and diffusely calcified. crowding of bronchovascular structures is present without overt pulmonary edema. patchy opacities in the lung bases likely reflect areas of atelectasis. no focal consolidation pleural effusion or pneumothorax is present. there are no acute osseous abnormalities. |
We present examples of generated reports and their corresponding ground truth from MIMIC-ABN and MIMIC-CXR. As shown in Table 3 and Table 4, our method (WCL) is able to generate similar contents which are aligned with the ground truth (GT) written by radiologists. For example, abnormal findings in specific positions (e.g., “low lung volumes” and “enlarged heart size”) are reported, and potential diseases are also noted (e.g., “atelectasis”).
5 Conclusion
In this paper, we present a weakly supervised contrastive learning framework for generating chest X-ray reports. Our contrastive loss could lead to better results on both clinical correctness and text generation metrics than previous methods. We also show that exposing the model to semantically-close negative samples improves generation performance. In the future, we will extend our method to other medical image datasets other than chest X-rays.
Acknowledgments
This work was partially supported by the Office of the Assistant Secretary of Defense for Health Affairs through the AIMM Research Program endorsed by the Department of Defense under Award No. W81XWH-20-1-0693 and NSF Award #1750063. Opinions, interpretations, conclusions and recommendations are those of the author and are not necessarily endorsed by the Department of Defense and the National Science Foundation.
References
- Anderson et al. (2018) Peter Anderson, Xiaodong He, Chris Buehler, Damien Teney, Mark Johnson, Stephen Gould, and Lei Zhang. 2018. Bottom-up and top-down attention for image captioning and visual question answering. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 6077–6086.
- Boag et al. (2020) William Boag, Tzu-Ming Harry Hsu, Matthew McDermott, Gabriela Berner, Emily Alesentzer, and Peter Szolovits. 2020. Baselines for chest x-ray report generation. In Machine Learning for Health Workshop, pages 126–140. PMLR.
- Chen et al. (2020a) Ting Chen, Simon Kornblith, Mohammad Norouzi, and Geoffrey Hinton. 2020a. A simple framework for contrastive learning of visual representations. In International conference on machine learning, pages 1597–1607. PMLR.
- Chen et al. (2020b) Zhihong Chen, Yan Song, Tsung-Hui Chang, and Xiang Wan. 2020b. Generating radiology reports via memory-driven transformer. arXiv preprint arXiv:2010.16056.
- Dai and Lin (2017) Bo Dai and Dahua Lin. 2017. Contrastive learning for image captioning. arXiv preprint arXiv:1710.02534.
- Denkowski and Lavie (2011) Michael Denkowski and Alon Lavie. 2011. Meteor 1.3: Automatic metric for reliable optimization and evaluation of machine translation systems. In Proceedings of the sixth workshop on statistical machine translation, pages 85–91.
- Gutmann and Hyvärinen (2010) Michael Gutmann and Aapo Hyvärinen. 2010. Noise-contrastive estimation: A new estimation principle for unnormalized statistical models. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, pages 297–304. JMLR Workshop and Conference Proceedings.
- Harzig et al. (2019) Philipp Harzig, Yan-Ying Chen, Francine Chen, and Rainer Lienhart. 2019. Addressing data bias problems for chest x-ray image report generation. arXiv preprint arXiv:1908.02123.
- He et al. (2020) Kaiming He, Haoqi Fan, Yuxin Wu, Saining Xie, and Ross Girshick. 2020. Momentum contrast for unsupervised visual representation learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pages 9729–9738.
- He et al. (2016) Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. 2016. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 770–778.
- Holtzman et al. (2019) Ari Holtzman, Jan Buys, Li Du, Maxwell Forbes, and Yejin Choi. 2019. The curious case of neural text degeneration. arXiv preprint arXiv:1904.09751.
- Huang et al. (2018) Jiaji Huang, Yi Li, Wei Ping, and Liang Huang. 2018. Large margin neural language model. arXiv preprint arXiv:1808.08987.
- Irvin et al. (2019) Jeremy Irvin, Pranav Rajpurkar, Michael Ko, Yifan Yu, Silviana Ciurea-Ilcus, Chris Chute, Henrik Marklund, Behzad Haghgoo, Robyn Ball, Katie Shpanskaya, et al. 2019. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 33, pages 590–597.
- Jing et al. (2020) Baoyu Jing, Zeya Wang, and Eric Xing. 2020. Show, describe and conclude: On exploiting the structure information of chest x-ray reports. arXiv preprint arXiv:2004.12274.
- Jing et al. (2017) Baoyu Jing, Pengtao Xie, and Eric Xing. 2017. On the automatic generation of medical imaging reports. arXiv preprint arXiv:1711.08195.
- Johnson et al. (2019) Alistair EW Johnson, Tom J Pollard, Nathaniel R Greenbaum, Matthew P Lungren, Chih-ying Deng, Yifan Peng, Zhiyong Lu, Roger G Mark, Seth J Berkowitz, and Steven Horng. 2019. Mimic-cxr-jpg, a large publicly available database of labeled chest radiographs. arXiv preprint arXiv:1901.07042.
- Khosla et al. (2020) Prannay Khosla, Piotr Teterwak, Chen Wang, Aaron Sarna, Yonglong Tian, Phillip Isola, Aaron Maschinot, Ce Liu, and Dilip Krishnan. 2020. Supervised contrastive learning. arXiv preprint arXiv:2004.11362.
- Lee et al. (2020) Seanie Lee, Dong Bok Lee, and Sung Ju Hwang. 2020. Contrastive learning with adversarial perturbations for conditional text generation. arXiv preprint arXiv:2012.07280.
- Li et al. (2019) Christy Y Li, Xiaodan Liang, Zhiting Hu, and Eric P Xing. 2019. Knowledge-driven encode, retrieve, paraphrase for medical image report generation. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 33, pages 6666–6673.
- Li et al. (2018) Yuan Li, Xiaodan Liang, Zhiting Hu, and Eric P Xing. 2018. Hybrid retrieval-generation reinforced agent for medical image report generation. In Advances in neural information processing systems, pages 1530–1540.
- Lin (2004) Chin-Yew Lin. 2004. Rouge: A package for automatic evaluation of summaries. In Text summarization branches out, pages 74–81.
- Lin et al. (2014) Tsung-Yi Lin, Michael Maire, Serge Belongie, James Hays, Pietro Perona, Deva Ramanan, Piotr Dollár, and C Lawrence Zitnick. 2014. Microsoft coco: Common objects in context. In European conference on computer vision, pages 740–755. Springer.
- Liu et al. (2019) Guanxiong Liu, Tzu-Ming Harry Hsu, Matthew McDermott, Willie Boag, Wei-Hung Weng, Peter Szolovits, and Marzyeh Ghassemi. 2019. Clinically accurate chest x-ray report generation. In Machine Learning for Healthcare Conference, pages 249–269. PMLR.
- Liu and Sun (2015) Yang Liu and Maosong Sun. 2015. Contrastive unsupervised word alignment with non-local features. In Proceedings of the AAAI Conference on Artificial Intelligence, volume 29.
- Nair and Hinton (2010) Vinod Nair and Geoffrey E Hinton. 2010. Rectified linear units improve restricted boltzmann machines. In Icml.
- Ni et al. (2020) Jianmo Ni, Chun-Nan Hsu, Amilcare Gentili, and Julian McAuley. 2020. Learning visual-semantic embeddings for reporting abnormal findings on chest x-rays. arXiv preprint arXiv:2010.02467.
- Oord et al. (2018) Aaron van den Oord, Yazhe Li, and Oriol Vinyals. 2018. Representation learning with contrastive predictive coding. arXiv preprint arXiv:1807.03748.
- Papineni et al. (2002) Kishore Papineni, Salim Roukos, Todd Ward, and Wei-Jing Zhu. 2002. Bleu: a method for automatic evaluation of machine translation. In Proceedings of the 40th annual meeting of the Association for Computational Linguistics, pages 311–318.
- Ranzato et al. (2015) Marc’Aurelio Ranzato, Sumit Chopra, Michael Auli, and Wojciech Zaremba. 2015. Sequence level training with recurrent neural networks. arXiv preprint arXiv:1511.06732.
- Smit et al. (2020) Akshay Smit, Saahil Jain, Pranav Rajpurkar, Anuj Pareek, Andrew Y Ng, and Matthew P Lungren. 2020. Chexbert: combining automatic labelers and expert annotations for accurate radiology report labeling using bert. arXiv preprint arXiv:2004.09167.
- Vinyals et al. (2015) Oriol Vinyals, Alexander Toshev, Samy Bengio, and Dumitru Erhan. 2015. Show and tell: A neural image caption generator. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 3156–3164.
- Xu et al. (2015) Kelvin Xu, Jimmy Ba, Ryan Kiros, Kyunghyun Cho, Aaron Courville, Ruslan Salakhudinov, Rich Zemel, and Yoshua Bengio. 2015. Show, attend and tell: Neural image caption generation with visual attention. In International conference on machine learning, pages 2048–2057.
- Yang et al. (2019) Zonghan Yang, Yong Cheng, Yang Liu, and Maosong Sun. 2019. Reducing word omission errors in neural machine translation: A contrastive learning approach.