Two-sample test based on maximum variance discrepancy
Abstract
In this article, we introduce a novel discrepancy called the maximum variance discrepancy for the purpose of measuring the difference between two distributions in Hilbert spaces that cannot be found via the maximum mean discrepancy. We also propose a two-sample goodness of fit test based on this discrepancy. We obtain the asymptotic null distribution of this two-sample test, which provides an efficient approximation method for the null distribution of the test.
1 Introduction
For probability distributions and , the test for the null hypothesis against an alternative hypothesis based on data and is known as a two-sample test. Such tests have applications in various areas. There is a huge body of literature on two-sample tests in Euclidean space, so we will not attempt a complete bibliography. In [6], a two-sample test based on Maximum Mean Discrepancy (MMD) is proposed, where the MMD is defined by (1) in Section 2. The MMD for a reproducing kernel Hilbert space associated with a positive definite kernel is defined as (2) in Section 2.
In this paper, we propose a novel discrepancy between two distributions defined as
and we call this the Maximum Variance Discrepancy (MVD), where is a covariance operator in . The MVD is composed by replacing the kernel mean embedding in (2) with a covariance operator; hence, it is natural to consider a two-sample test based on the MVD. A related work can be found in [4], where a test for the equality of covariance operators in Hilbert spaces was proposed.
Our aim in this research is to clarify the properties of the MVD test from two perspectives: an asymptotic investigation as , and its practical implementation. We first obtain the asymptotic distribution of a consistent estimator of , under . We also derive the asymptotic distribution of under the alternative hypothesis . Furthermore, we consider a sequence of local alternative distributions for and address the asymptotic distribution of under this sequence. For practical purposes, a method to approximate the distribution of the test by under is developed. The method is based on the eigenvalues of the centered Gram matrices associated with the dataset. Those eigenvalues will be shown to be estimators of the weights appearing in the asymptotic null distribution of the test. Hence, the method based on the eigenvalues is expected to provide a fine approximation of the distribution of the test. However, this approximation does not actually work well. Therefore, we further modify the method based on the eigenvalues, and the obtained method provides a better approximation.
The rest of this paper is structured as follows. Section 2 introduces the framework of the two-sample test and defines the test statistics based on the MVD. In addition, the representation of test statistics based on the centered Gram matrices is described. Section 3.1 develops the asymptotics for the test by the MVD under . The test by the MVD under is addressed in Section 3.2. Furthermore, the behavior of the test by the MVD under the local alternative hypothesis is clarified in Section 3.3. Section 3.4 describes the estimation of the weights that appear in the asymptotic null distribution obtained in Section 3.1. Section 4 examines the implementation of the MVD test with a Gaussian kernel in the Hilbert space . Section 4.1 introduces the modification of the approximate distribution given in Section 3.4. Section 4.2 reports the results of simulations for the type I error and the power of the MVD and MMD tests. Section 5 presents the results of applications to real data sets, including high-dimension low-sample-size data. Conclusions are given in Section 6. All proofs of theoretical results are provided in Section 7.
2 Maximum Variance Discrepancy
Let be a separable Hilbert space and be a measurable space. Let be the inner product of and be the associated norm. Let and denote a sample of independent and identically distributed (i.i.d.) random variables drawn from unknown distributions and , respectively. Our goal is to test whether the unknown distributions and are equal.
Let us define the null hypothesis and the alternative hypothesis . Following [6], the gap between two distributions and on is measured by:
| (1) |
where is a class of real-valued functions on . Regardless of , always defines a pseudo-metric on the space of probability distributions. Let be the unit ball of a reproducing kernel Hilbert space associated with a characteristic kernel (see [2] and [5] for details) and assume that and . Then, the MMD in is defined as the distance between and as follows:
| (2) |
where and are called kernel mean embeddings of and , respectively, in (see [6]). The MMD focuses on the difference between distributions and depending on the difference between the means of and in . The motivation for this research is to focus on the difference between the distributions and due to the difference between those variances in , based on a similar idea as the MMD. Assume and , then the variance is defined by
Here, for any , the tensor product is defined as the operator , is defined as , and (see Section II.4 in [12] for details). Let and . Then, we define the MVD in as
which can be seen as a discrepancy between distributions and . The can be estimated by
| (3) |
where
and
Let the Gram matrices be , , and ; the centering matrix be ; and the centered Gram matrices be , , and , where is the identity matrix. This test statistic can be expanded as:
We investigate the and when , the kernel is the Gaussian kernel:
| (4) |
, and . Under this setting, straightforward calculations using the properties of Gaussian density yield the following result for MMD:
Proposition 1.
When and is the Gaussian kernel in (4), we have
The result for MVD is also obtained as follows using the Gaussian density property as well as the result for MMD:
Proposition 2.
When and is the Gaussian kernel in (4), we have
Corollary 1.
We investigate the behavior of and for the difference of from (0,1) by using Corollary 1. A sensitive reaction to the difference of from (0,1) means that it can sensitively react to differences between distributions from . Using such a discrepancy in the framework of the test is expected to correctly reject under .
More generally, kernel based on a constant and a positive definite kernel is also positive definite. Then, and calculated by the kernel hold and for any distributions and using and calculated by the kernel .
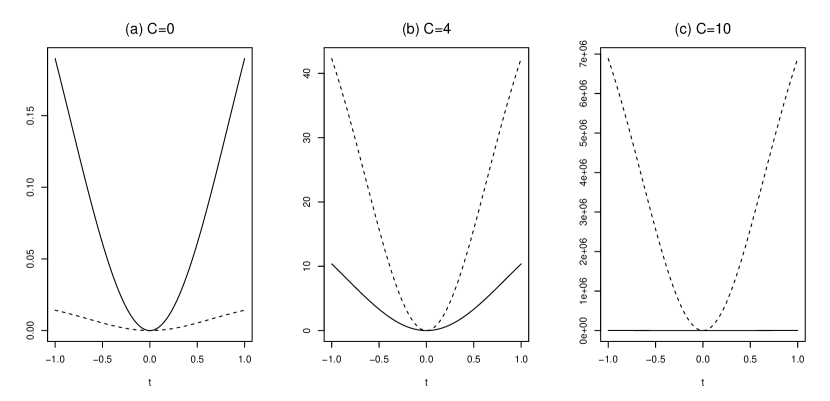
The graph of and is displayed for each when in Figure 1 and for each when in Figure 2. The kernel is a Gaussian kernel in (4), and the parameters are , , and in both Figures 1 and 2. Figure 1 shows the and for the difference of the mean from the standard normal distribution. In Figure 1 (a), is larger than , but in Figures 1 (b) and (c), where the value of is increased, is larger than for each . In addition, Figure 2 shows the reaction of the MMD and MVD to the difference of the covariance matrix from the standard normal distribution, and is larger than for each when is large. This means that MVD is more sensitive to differences from the standard normal distribution than MMD for with large . The fact that there is a kernel for which MVD is larger than MMD is a motivation for the two-sample test based on MVD in the next section.

3 Test statistic for two-sample problem
We consider a two-sample test based on for and , and is defined as a test statistic. If is large, then the null hypothesis is rejected since is the difference between and . The condition to derive the asymptotic distribution of this test statistic is as follows:
Condition.
, and .
3.1 Asymptotic null distribution
In this section, we derive an asymptotic distribution of under . In what follows, the symbol designates convergence in distribution.
Theorem 1.
Suppose that Condition is satisfied. Then, under , as ,
where and is an eigenvalue of .
It is generally not easy to utilize such an asymptotic null distribution because it is an infinite sum and determining the weights included in the asymptotic distribution is itself a difficult problem. For practical purposes, a method to approximate the distribution of the test by under is developed in Section 4. The method is based on the eigenvalues of the centered Gram matrices associated with the data set in Section 3.4.
3.2 Asymptotic non-null distribution
In this section, an asymptotic distribution of under is investigated.
Theorem 2.
Suppose that Condition is satisfied. Then, under , as ,
where
and
Remark 1.
We see by Theorem 2 that
where . Thus, we can evaluate the power of the test by as
as , where is the -quantile of the distribution of under , and is the distribution function of the standard normal distribution. Therefore, this test is consistent.
3.3 Asymptotic distribution under contiguous alternatives
In this section, we develop an asymptotic distribution of under a sequence of local alternative distributions for .
Theorem 3.
Let and . Suppose that Condition is satisfied. Let be a kernel defined as
| (5) |
and
and let be a self-adjoint operator defined as
| (6) |
(see Sections VI.1, VI.3, and VI.6 in [12] for details). Then, as
where ,
and and are, respectively, the eigenvalue of and the eigenfunction corresponding to .
The following proposition claims that the eigenvalues appearing in Theorem 3 are the same as the eigenvalues appearing in Theorem 1:
From Theorems 1 and 3 and Proposition 3, it can be seen that the local power of the test by is dominated by the noncentrality parameters. It follows that
by which we obtain
In addition, from Theorem 1 in [10], we have
Hence, the local power reveals not only the difference between and but also that between and .
3.4 Null distribution estimates using Gram matrix spectrum
The asymptotic null distribution was obtained in Theorem 1, but it is difficult to derive its weights. The following theorem shows that this weight can be estimated using the estimator of .
Theorem 4.
Assume that . Let and be the eigenvalues of and , respectively, where
Then, as
where .
In addition, Proposition 4 claims the eigenvalues of and the Gram matrix are the same.
Proposition 4.
The Gram matrix is defined as
Then, the eigenvalues of and are the same.
Remark 2.
By Proposition 4, the critical value can be obtained by calculating using the eigenvalue of . In addition, the matrix is expressed as
| (7) |
where is the Hadamard product. The Gram matrix is a positive definite, but has eigenvalue 0 since .
4 Implementation
This section proposes corrections to the asymptotic distribution for both the MVD and MMD tests, and describes the results of simulations of the type-I error and power for the modifications. The MMD test is a two-sample test for and using the test statistic:
The of the asymptotic null distribution is the infinite sum of the weighted chi-square distribution, which is the same as in (3). The approximate distribution can be obtained by estimating the eigenvalues by the centered Gram matrix (see [7] for details).
4.1 Approximation of the null distribution
In this section, we discuss methods to approximate the null distributions of the MVD and MMD tests. The asymptotic null distribution of the MVD test was obtained in Theorem 1 as an infinite sum of weighted chi-squared random variables with one degree of freedom, and according to Theorem 4, those weights can be estimated by the eigenvalues of the matrix . Similar results was obtained for MMD by [7]. However, this approximate distribution based on this estimated eigenvalue does not actually work well. In fact, by comparing the simulated exact null distribution with this approximate distribution based on estimated eigenvalues, it can be seen that variance of the approximate distribution is larger than that of the simulated exact null distribution. We modify the approximate distribution by obtaining the variance of this simulated exact null distribution. The variance of the exact null distribution is obtained as the following proposition:
Proposition 5.
Assume that . Then under ,
Proposition 5 leads to
| (8) |
with respect to and . If we can estimate the variance at and that is less than and , respectively, we can estimate by using (8). In addition, the method of estimating by choosing from without replacement is known as subsampling.
The following proposition for MMD shows a similar result to MVD:
Proposition 6.
Assume that . Then, under
4.1.1 Subsampling method
We used the subsampling method to estimate and (see Section 2.2 in [11] for details). In order to obtain two samples under the null hypothesis, we divide into and . Then, we randomly select and from and without replacement, which repaet in each iteration . These randomly selected values generate the replications of the test statistic
for iterations . The generated test statistics in iterations estimate by calculating the unbiased sample variance:
where . According to (8), is estimated by
| (9) |
We also estimate by using
| (10) |
from Proposition 6, where
for ,
and .
The columns of in Table 1 and in Table 2 are variances of and , which are estimated by a simulation of 10,000 iterations with and for each and . The subsampling variances in (9) and in (10) with are given in the columns labeled “Subsampling” for . Tables 1 and 2 show that subsampling variances and estimate the exact variances well. However, these variances tend to underestimate the exact variances.
We investigate how much smaller is than the variance of by performing a linear regression of and the variance of with an intercept equal to 0, with the same for the MMD. The results are shown in Figure 3; (a) and (b) show results for the MVD and (c) and (d) show results for the MMD, which are respectively and and and cases. In Figure 3, the axis is or and the axis is variance of or for each and in Table 1 or Table 2. The line in Figure 3 is a regression line found by the least-squares method in the form . It can be seen from Figure 3 that when and are multiplied by the term associated regression coefficient, they approach the variances and . The coefficient of linear regression with intercept 0 is written in the row labeled “slope of the line” in Tables 1 and 2.
| Subsampling | ||||||
| 5 | (200,200) | 0.06880 | 0.04341 | 0.05168 | 0.04902 | |
| 5 | (500,500) | 0.06881 | 0.03821 | 0.04897 | 0.04921 | |
| 5 | (200,200) | 0.07254 | 0.04246 | 0.05138 | 0.05798 | |
| 5 | (500,500) | 0.07188 | 0.04052 | 0.05500 | 0.05593 | |
| 10 | (200,200) | 0.00850 | 0.00602 | 0.00812 | 0.00898 | |
| 10 | (500,500) | 0.00845 | 0.00674 | 0.00751 | 0.00753 | |
| 10 | (200,200) | 0.01280 | 0.00937 | 0.01224 | 0.01377 | |
| 10 | (500,500) | 0.01270 | 0.01032 | 0.01251 | 0.01255 | |
| 20 | (200,200) | 0.00049 | 0.00048 | 0.00070 | 0.00094 | |
| 20 | (500,500) | 0.00043 | 0.00031 | 0.00046 | 0.00060 | |
| 20 | (200,200) | 0.00166 | 0.00152 | 0.00261 | 0.00330 | |
| 20 | (500,500) | 0.00147 | 0.00122 | 0.00165 | 0.00204 | |
| (200,200) | 1 | 1.63621 | 1.35769 | 1.29601 | ||
| slope of the line | (500,500) | 1 | 1.76057 | 1.33845 | 1.3232 | |
| both | 1 | 1.69348 | 1.34798 | 1.30928 | ||
| Subsampling | ||||||
| 5 | (200,200) | 0.57100 | 0.47044 | 0.50315 | 0.57047 | |
| 5 | (500,500) | 0.66068 | 0.54518 | 0.63054 | 0.58848 | |
| 5 | (200,200) | 0.65987 | 0.51867 | 0.57258 | 0.54567 | |
| 5 | (500,500) | 0.75903 | 0.63563 | 0.68024 | 0.65349 | |
| 10 | (200,200) | 0.16205 | 0.09213 | 0.12017 | 0.12940 | |
| 10 | (500,500) | 0.16279 | 0.16106 | 0.16809 | 0.18334 | |
| 10 | (200,200) | 0.25656 | 0.14104 | 0.18435 | 0.20457 | |
| 10 | (500,500) | 0.25836 | 0.24670 | 0.25567 | 0.26402 | |
| 20 | (200,200) | 0.02757 | 0.02135 | 0.02632 | 0.02814 | |
| 20 | (500,500) | 0.02784 | 0.02255 | 0.02407 | 0.02615 | |
| 20 | (200,200) | 0.07642 | 0.07404 | 0.08121 | 0.08744 | |
| 20 | (500,500) | 0.07856 | 0.05714 | 0.07492 | 0.07320 | |
| (200,200) | 1 | 1.27369 | 1.16037 | 1.11083 | ||
| slope of the line | (500,500) | 1 | 1.18426 | 1.07578 | 1.12080 | |
| both | 1 | 1.21990 | 1.10951 | 1.11643 | ||




Remark 3.
Since the subsampling variance is an unbiased sample variance of times, we get
However, it is not easy to estimate . This fact caused us to modify subsampling variances in (9) and in (10) to and using , which is determined from the regression coefficient in the row “slope of the line” in Tables 1 and 2.
4.1.2 Modification of approximation distribution
Since the subsampling variance underestimates , as seen in Table 1 and 2, we use positive to estimate by in (9). This underestimation is the same for the MMD test and is estimated by in (10) with positive . Our approximation of the null distribution is based on a modification of the large variance of to . The method aims to approximate the exact null distribution by using
| (11) |
The parameters and are determined so that the means of and are equal and the variance of is equal to , which can be established by
and
This approximation method can be similarly discussed for the MMD test using . In this paper, the parameter is determined using the value of “slope of the line” in Tables 1 and 2. Figure 4 shows that this can approximate the simulated exact distribution better than . Algorithm shows how to obtain the critical value of the MVD test using this modification. The algorithm for the MMD test can be obtained by changing and in Algorithm to and .


4.2 Simulations
In this section, we investigate the performance of under a specific null hypothesis and specific alternative hypotheses when and is the Gaussian kernel in (4). In particular, a Monte Carlo simulation is performed to observe the type-I error and the power of the MVD and MMD tests. Two cases are implemented: a uniform distribution and an exponential distribution , with , all of which have means and variances 0 and 1, respectively. The critical values are determined based on in Section 4.1.2 from a normal distribution. The type-I error of can be obtained by counting the number of times exceeds the critical value in 1,000 iterations under the null hypothesis. Next, the estimated power of is similarly obtained by counting the number of times exceeds the critical value under each alternative distribution in 1,000 iterations. We execute the above for and and , and . It is known that the selection of the value of involved in the Gaussian kernel affects the performance. We utilize depending on dimension . The significance level is . The critical values are determined on the basis of in Section 4.1.2 from a normal distribution. The type-I error and estimated power can be obtained by counting how many times exceeds the critical values in 1,000 iterations under and . With and , for MVD is and for MMD is by “slope of the line” in Tables 1 and 2. The following can be seen from Table 3:
-
•
Table 3 shows that the probabilities of type-I error at and 10 are near the significance level of for both the MVD and MMD.
-
•
The probability of type-I error at exceeds the significance level of for the MVD, but decreases as increases.
-
•
It can be seen that the critical value by of the MVD tends to be estimated as less than that point of the null distribution.
-
•
In hypothesis , it can be seen that the MVD has a higher power than the MMD.
-
•
It can be seen that the MVD and MMD have higher powers for hypothesis than hypothesis and it is difficult to distinguish between the normal distribution and the uniform distribution by the MVD and MMD for a Gaussian kernel.
-
•
Note that the critical value changes depending on the distribution of the null hypothesis.
| MVD | MMD | |||||||
|---|---|---|---|---|---|---|---|---|
| Type-I error | Type-I error | |||||||
| 5 | (200,200) | 0.060 | 0.797 | 1 | 0.047 | 0.401 | 1 | |
| 5 | (500,500) | 0.072 | 1 | 1 | 0.063 | 0.877 | 1 | |
| 5 | (200,200) | 0.056 | 0.728 | 1 | 0.052 | 0.305 | 1 | |
| 5 | (500,500) | 0.067 | 0.999 | 1 | 0.053 | 0.735 | 1 | |
| 10 | (200,200) | 0.073 | 0.612 | 1 | 0.086 | 0.342 | 1 | |
| 10 | (500,500) | 0.047 | 0.991 | 1 | 0.040 | 0.630 | 1 | |
| 10 | (200,200) | 0.054 | 0.482 | 1 | 0.086 | 0.235 | 1 | |
| 10 | (500,500) | 0.034 | 0.955 | 1 | 0.044 | 0.363 | 1 | |
| 20 | (200,200) | 0.279 | 0.816 | 1 | 0.082 | 0.239 | 1 | |
| 20 | (500,500) | 0.099 | 0.948 | 1 | 0.068 | 0.477 | 1 | |
| 20 | (200,200) | 0.060 | 0.332 | 1 | 0.047 | 0.113 | 0.989 | |
| 20 | (500,500) | 0.034 | 0.728 | 1 | 0.069 | 0.240 | 1 | |
5 Application to real datasets
The MVD test was applied to some real data sets. The significance level was and the critical value was obtained through 10,000 iterations of based on the eigenvalues of the matrix . We also calculated the critical value of the approximate distribution according to Algorithm in Section 4.1.2. The calculates the critical value for the MMD test from the distribution obtained based on Theorem 1 in [7] through 10,000 iterations.
5.1 USPS data
The USPS dataset consists of handwritten digits represented by a grayscale matrix (https://www.csie.ntu.edu.tw/~cjlin/libsvmtools/datasets/multiclass.html#usps). The sizes of each sample are shown in Table 4.
| index | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|---|
| sample size | 359 | 264 | 198 | 166 | 200 | 160 | 170 | 147 | 166 | 177 |
Each group was divided into two sets of sample size 70 and the MVD test was applied to each set. Table 5 shows the results of applying the MVD and MMD tests to this USPS dataset. Parameters , , and are adopted and for the MVD and for the MMD were utilized from the slope of the line in Tables 1 and 2. In each cell, the values of and for each number are written, with the values of in parentheses.
From Table 5, there is a tendency that different groups will be rejected and that the same groups are not rejected by the MVD test. For USPS 2 and USPS 2, the value of is 2.953, which is larger than but smaller than . On the other hand, for the MMD test, the value of is 5.014, which is larger than both and . By modifying the distribution, there is a tendency to reject the null hypothesis.
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 3.056 | 0.680 | 3.416 | 2.742 | 2.747 | 3.319 | 2.705 | 2.079 | 2.884 | 1.941 | |
| 2.755 | 0.572 | 2.940 | 2.317 | 2.349 | 2.785 | 2.336 | 1.823 | 2.431 | 1.719 | |
| 0 | 2.241 | 6.328 | 6.803 | 6.834 | 7.390 | 6.589 | 7.117 | 7.513 | 6.930 | 4.573 |
| (2.740) | ||||||||||
| 1 | (124.6) | 0.287 | 4.269 | 4.290 | 4.356 | 4.393 | 5.132 | 4.265 | 4.233 | 4.170 |
| (0.585) | ||||||||||
| 2 | (34.38) | (94.14) | 2.953 | 4.730 | 5.253 | 5.053 | 5.880 | 5.264 | 4.732 | 5.165 |
| (5.014) | ||||||||||
| 3 | (42.61) | (105.0) | (26.78) | 2.383 | 5.248 | 4.239 | 6.022 | 5.242 | 4.575 | 5.022 |
| (3.345) | ||||||||||
| 4 | (55.81) | (93.27) | (36.46) | (45.34) | 2.067 | 5.259 | 6.237 | 4.930 | 4.849 | 3.717 |
| (2.745) | ||||||||||
| 5 | (30.65) | (95.83) | (24.64) | (18.91) | (35.32) | 2.761 | 5.757 | 5.434 | 4.814 | 5.106 |
| (3.822) | ||||||||||
| 6 | (39.15) | (102.6) | (30.11) | (47.36) | (48.80) | (29.49) | 2.261 | 6.527 | 5.946 | 6.344 |
| (5.643) | ||||||||||
| 7 | (72.41) | (111.3) | (50.29) | (52.91) | (45.54) | (51.29) | (74.30) | 1.560 | 5.142 | 4.062 |
| (1.785) | ||||||||||
| 8 | (44.46) | (86.90) | (25.20) | (28.77) | (31.13) | (25.01) | (40.92) | (51.27) | 2.055 | 4.352 |
| (2.666) | ||||||||||
| 9 | (71.81) | (95.38) | (51.48) | (49.58) | (25.87) | (46.76) | (70.81) | (31.19) | (33.73) | 1.677 |
| (2.336) | ||||||||||
| (4.983) | (2.191) | (4.698) | (4.352) | (4.343) | (4.691) | (4.550) | (3.917) | (4.424) | (3.767) | |
| (5.228) | (1.749) | (4.502) | (3.928) | (3.961) | (4.085) | (4.510) | (4.104) | (4.245) | (4.455) |
5.2 MNIST data
The MNIST dataset consists of images of pixels in size (http://yann.lecun.com/exdb/mnist). The sizes of each sample are shown in Table 6.
| index | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|---|
| sample size | 5,923 | 6,742 | 5,958 | 6,131 | 5,842 | 5,421 | 5,918 | 6,265 | 5,851 | 5,949 |
The MNIST data are divided into two sets of sample size 2,000 and the MVD and MMD tests are applied. Table 7 shows the results of applying the MVD and MMD tests to the MNIST data. The approximate distribution is calculated with , , , and . As in Table 5, the values of and are written in each cell, with the values of in parentheses. In Table 7, tends to take a larger value than both and . This result is the same for the MMD test. The MVD and MMD tests tend to reject the null hypothesis with the modifications in Section 4.1.2.
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 4.194 | 3.883 | 4.202 | 4.196 | 4.189 | 4.195 | 4.182 | 4.163 | 4.197 | 4.171 | |
| 3.993 | 3.937 | 3.993 | 3.989 | 3.990 | 3.993 | 3.989 | 3.990 | 3.989 | 3.988 | |
| 0 | 3.999 | 34.86 | 4.207 | 4.268 | 4.394 | 4.304 | 4.599 | 5.240 | 4.256 | 4.861 |
| (4.092) | ||||||||||
| 1 | (217.4) | 5.379 | 34.68 | 34.75 | 34.86 | 34.80 | 35.08 | 35.66 | 34.73 | 35.33 |
| (15.42) | ||||||||||
| 2 | (10.77) | (210.5) | 4.001 | 4.118 | 4.245 | 4.156 | 4.451 | 5.088 | 4.106 | 4.711 |
| (4.131) | ||||||||||
| 3 | (13.55) | (213.4) | (9,806) | 4.007 | 4.306 | 4.211 | 4.512 | 5.146 | 4.164 | 4.769 |
| (4.208) | ||||||||||
| 4 | (16.56) | (216.1) | (12.83) | (15.59) | 4.020 | 4.341 | 4.637 | 5.262 | 4.292 | 4.805 |
| (4.482) | ||||||||||
| 5 | (13.35) | (213.9) | (10.10) | (11.57) | (15.12) | 4.018 | 4.546 | 5.188 | 4.201 | 4.806 |
| (4.357) | ||||||||||
| 6 | (19.39) | (219.5) | (16.06) | (18.90) | (21.28) | (18.29) | 4.031 | 5.485 | 4.499 | 5.105 |
| (4.573) | ||||||||||
| 7 | (28.85) | (225.2) | (24.85) | (27.49) | (28.28) | (27.56) | (34.24) | 4.067 | 5.138 | 5.625 |
| (4.625) | ||||||||||
| 8 | (13.12) | (211.3) | (9.261) | (11.37) | (14.72) | (11.51) | (18.19) | (26.97) | 4.005 | 4.755 |
| (4.190) | ||||||||||
| 9 | (24.80) | (223.2) | (21.11) | (23.23) | (19.05) | (22.80) | (29.84) | (29.91) | (22.07) | 4.046 |
| (4.686) | ||||||||||
| (4.210) | (4.718) | (4.208) | (4.206) | (4.210) | (4.209) | (4.218) | (4.238) | (4.207) | (4.224) | |
| (4.058) | (5.137) | (4.024) | (4.038) | (4.077) | (4.107) | (4.092) | (4.137) | (4.039) | (4.159) |
5.3 Colon data
The Colon dataset contains gene expression data from DNA microarray experiments of colon tissue samples with = 2,000 and (see [1] for details). Among the 62 samples, 40 are tumor tissues and 22 are normal tissues. Tables 8 and 9 show the results of the MVD and MMD tests for tumor and normal. The “tumor vs. normal” column shows the values of and for tumor and normal. The “normal” and “tumor” columns show, and calculated from respectively the normal tissues and tumor tissues datasets.
For the MVD, does not exceed , but exceeds by modifying the approximate distribution. By contrast, for the MMD, exceeds both and without modifying the approximate distribution.
| normal | tumor | ||||
|---|---|---|---|---|---|
| tumor vs. normal | |||||
| 3.867 | 3.536 | 5.280 | 3.728 | 5.050 | |
| 2.291 | 2.097 | 2.907 | 2.258 | 2.846 | |
| 0.684 | 0.660 | 0.879 | 0.757 | 0.906 | |
| normal | tumor | ||||
|---|---|---|---|---|---|
| tumor vs. normal | |||||
| 6.695 | 4.456 | 6.201 | 4.618 | 5.713 | |
| 8.787 | 3.974 | 4.827 | 3.945 | 4.491 | |
| 6.282 | 2.439 | 2.754 | 2.412 | 2.634 | |
Next, tumor was divided into tumor 1 and tumor 2 , and two-sample tests by the MVD and MMD were applied. The results are shown in Tables 10 and 11, with the values for and in the column “tumor 1 vs. tumor 2”. In Table 10, when , exceeds , but in other cases does not exceed and does not exceed and . Table 11 shows that, for all , exceeds for the MMD test, but does not exceed .
| tumor 1 | tumor 2 | ||||
|---|---|---|---|---|---|
| tumor 1 vs. tumor 2 | |||||
| 3.379 | 3.180 | 4.596 | 3.245 | 4.942 | |
| 1.858 | 1.915 | 2.502 | 2.085 | 2.875 | |
| 0.558 | 0.629 | 0.800 | 0.727 | 0.921 | |
| tumor 1 | tumor 2 | ||||
|---|---|---|---|---|---|
| tumor 1 vs. tumor 2 | |||||
| 4.627 | 4.064 | 5.621 | 3.961 | 5.782 | |
| 4.123 | 3.305 | 4.206 | 3.426 | 4.551 | |
| 2.453 | 1.942 | 2.377 | 2.102 | 2.656 | |
6 Conclusion
We defined a Maximum Variance Discrepancy (MVD) with a similar idea to the Maximum Mean Discrepnacy (MMD) in Section 2. We derived the asymptotic null distribution for the MVD test in Section 3.1. This was the infinite sum of the weighted chi-square distributions. In Section 3.2, we derived an asymptotic nonnull distribution for the MVD test, which was a normal distribution. The asymptotic normality of the test under the alternative hypothesis showed that the two-sample test by the MVD has consistency. Furthermore, we developed an asymptotic distribution for the test under a sequence of local alternatives in Section 3.3. This was the infinite sum of weighted noncentral chi-squared distributions. We constructed an estimator of asymptotic null distributed weights based on the Gram matrix in Section 3.4. The approximate distribution of the null distribution by these estimated weights does not work well, so we modified it in Section 4.1. In the simulation of the power reported, we found that the power of the two-sample test by the MVD was larger than that of the MMD. We confirmed in Section 5 that the two-sample test by the MVD works for real data-sets.
7 Proofs
Lemma 1.
Suppose that Condition is satisfied. Then, as ,
where
Lemma 2.
Let . Suppose that Condition is satisfied. Then, as , following evaluates are obtained
-
(i)
-
(ii)
-
(iii)
Lemma 3.
Let and . Suppose that Condition is satisfied. Then, as ,
7.1 Proof of Proposition 1
The kernel mean embedding with the Gaussian kernel in (4) is obtained
| (12) |
and the norm of that is derived
| (13) |
by Proposition 4.2 in [9]. We use the property of Gaussian density
| (14) |
where
and designates the density of , see e.g. Appendix C in [13]. The property of Gaussian density (14) is used repeatedly to calculate , and we get
| (15) |
where
Using this results (13) and (15), is obtained as
7.2 Proof of Proposition 2
In the following proof, the when using the Gaussian kernel in (4) is calculated by repeatedly using (14). From the expansion of the norm
| (16) |
it is sufficient for us to calculate . The definition of and the tensor product lead to
| (17) |
We calculate each of these terms. The first term is derived as
| (18) |
by repeatedly using (14). By using the expression of kernel mean embedding in (12) and the property of (14), we obtain the second term
| (19) |
The third term is derived
| (20) |
by the same calculation as . Finally, the fourth term is calculated as follows
| (21) |
by using (15). Hence, combining (17) and (18)-(21) yields
| (22) |
The following results are obtained by using (22):
| (23) | |||
| (24) |
and
| (25) |
Therefore, we give
7.3 Proof of Theorem 1
7.4 Proof of Theorem 2
By Lemma 1, it suffices to derive the asymptotic distribution of
Let us expand the following quantity
which converges in distribution to by the central limit theorem.
7.5 Proof of Lemma 1
A direct calculation gives
From direct expansion , we have
as .
7.6 Proof of Theorem 3
From Lemma 3 it is sufficient for us to derive the asymptotic distribution of . It follows from direct calculations that
| (26) |
where
In addition, we see that
| (27) |
by (i) in Lemma 2. Thus, (27) leads that
| (28) |
Further, following result is obtained by (i) and (ii) in Lemma 2
| (29) |
These results (28) and (29) yield that
| (30) |
By using (28) and (30) to (26), we obtain
where is in (5). Since , in (6) is a Hilbert–Schmidt operator by Theorem VI.22 in [12]. Therefore, from Theorem 1 in [10], we have
where is eigenvalue of and is eigenfunction corresponding to , each satisfies
with Kronecker’s delta. From , we have
and
Since direct calculation, we get
and
Hence
and
where
Therefore, by using the central limit theorem for s, we get
as .
7.7 Proof of Lemma 2
(i) First, we prove . For all , there exists such that
for all . In addition, there exists such that
for all . We put
and . Since
for all , we get
Therefore, we obtain .
(ii) Next, we prove . For all , we put
and
Then, there exists such that
for all . In addition, there exists such that
for all . and there exists such that
for all .
Let . For all , we obtain that
Therefore, is proved.
(iii) Finally, we prove . We get
by a expansion of . Using (i) and (ii) leads to the following:
7.8 Proof of Lemma 3
7.9 Proof of Proposition 3
Let be the operator with spectral representation , and be the operator
for all and . We consider the adjoint operator of this ,
for all and , hence we get that adjoint operator of is
for all . Furthermore, since
for all , is the identity operator from to . It follows from direct calculations for that
for all an , thus we see that . Therefore, and are the eigenvalue and eigenfunction of , by the following that
and is an orthonormal system in which holds
for all . In fact,
shows that and are eigenvalue and eigenfunction of .
7.10 Proof of Theorem 4
Let . Then
| (32) |
By the definition of trace of the Hilbert–Schmidt operator, we get the following inequality
| (33) |
By using a notation
we have
| (34) |
Since direct calculation, we get the following three expressions,
| (35) |
| (36) |
and
| (37) |
By (35), (36) and (37) are combined into (34), we have an expression
Therefore,
| (38) |
By the law of large number in Hilbert spaces (see [8]), we have
| (39) |
and we get
| (40) |
| (41) |
and
| (42) |
Next our focus goes to . We have by direct computations that
Since Condition is assumed, the followings hold
and
as by the law of large numbers. Hence, we get
| (43) |
Also, we see that
by Cauchy–Schwartz inequality, and we get
as . Hence, we get
| (44) |
since . By the same argument as for in (44), we have
| (45) |
The equations (39), (40), (41), (42), (43), (44) and (45) are combined into (38), which leads to
| (46) |
Therefore, we get
| (47) |
Next, we consider . By and are compact Hermitian operators and (28) of [3],
Futhermore, we have got , hence
| (48) |
as . Also,
| (49) |
and since by (47), we obtain
| (50) |
as . In addition, as by (47), we get
Finally, we shall show , as . From Chebyshev’s inequality, for any ,
as . Therefore, as .
7.11 Proof of Proposition 4
Let be a kernel defined as,
and the associated function represent for all . Then, is defined by
for any . The conjugate operator of (see Section VI.2 in [12] for details) is obtained as for all , because for all ,
Let and be the eigenvalue and eigenvector of , respectevely. Then, it is holds that
from the definition of eigenvalue and eigenvector. By mapping both sides with ,
Hence, the eigenvalues of are that of .
Conversely, let and be the eigenvalue and correspondent eigenvector of , then
and
form mapping both sides with , hence the eigenvalue of are that of .
7.12 Proof of Proposition 5
7.13 Proof of Proposition 6
Since
first, we need to calculate
From the expected values of each term are obtained as
we get
| (53) |
under .
7.14 Proof of (7)
The -th element of the matrix is
Each term of this can be expressed as
and
Therefore,
which gives the expression (7).
References
- [1] U. Alon, N. Barkai, D. A. Notterman, K. Gish, S. Ybarra, D. Mack, and A. J. Levine. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proceedings of the National Academy of Sciences, 96(12):6745–6750, 1999.
- [2] N. Aronszajn. Theory of reproducing kernels. Transactions of the American Mathematical Society, 68(3):337–404, 1950.
- [3] R. Bhatia and L. Elsner. The Hoffman-Wielandt inequality in infinite dimensions. Proceedings of the Indian Academy of Sciences - Mathematical Sciences, 104(3):483–494, 1994.
- [4] G. Boente, D. Rodriguez, and M. Sued. Testing equality between several populations covariance operators. Annals of the Institute of Statistical Mathematics, 70(4):919–950, 2018.
- [5] K. Fukumizu, A. Gretton, X. Sun, and B. Schölkopf. Kernel measures of conditional dependence. Advances in Neural Information Processing Systems 20 - Proceedings of the 2007 Conference, pages 1–13, 2009.
- [6] A. Gretton, K. M. Borgwardt, M. Rasch, B. Schölkopf, and A. J. Smola. A kernel method for the two-sample-problem. In B. Schölkopf, J. C. Platt, and T. Hoffman, editors, Advances in Neural Information Processing Systems, volume 19, pages 513–520. MIT Press, 2007.
- [7] A. Gretton, K. Fukumizu, Z. Harchaoui, and B. Sriperumbudur. A fast, consistent kernel two-sample test. Advances in Neural Information Processing Systems, pages 673–681, 2009.
- [8] J. Hoffmann-Jorgensen and G. Pisier. The law of large numbers and the central limit theorem in Banach spaces. The Annals of Probability, 4(4):587–599, 1976.
- [9] J. Kellner and A. Celisse. A one-sample test for normality with kernel methods. Bernoulli, 25(3):1816–1837, 2019.
- [10] H. Q. Minh, P. Niyogi, and Y. Yao. Mercer’s theorem, feature maps, and smoothing. In Proceedings of the 19th Annual Conference on Learning Theory, COLT’06, pages 154–168, Berlin, Heidelberg, 2006. Springer-Verlag.
- [11] D. Politis, D. Wolf, J. Romano, M. Wolf, P. Bickel, P. Diggle, and S. Fienberg. Subsampling. Springer Series in Statistics. Springer New York, 1999.
- [12] M. Reed and B. Simon. Functional Analysis. Methods of Modern Mathematical Physics. Elsevier Science, 1981.
- [13] M. P. Wand and M. C. Jones. Kernel Smoothing. Chapman & Hall, New York, 1994.