0
Supplementary Information
Supplementary Notes
Note 1: Averaged intermolecular angles
For an arbitrary solute molecule , we have
| (1) |
where is the intermolecular angle between characteristic vectors on molecule and molecule , and is a switching function of intermolecular distance . In order to eliminate the mirror image symmetry, the hyperbolic tangent switching function is implemented in Eq. 1. We then compute the mean of over the group of solutes to obtain the averaged intermolecular angles and , for which the subscripts refer to the specific intermolecular angles defined in the context of pair orientational entropy. and are computed as the second moments of the and distributions.
Note 2: Coordination number
We calculate the coordination number of urea molecules by the following continuous and differentiable expression,
| (2) |
where is the distance between reference sites of molecules and , and is the cutoff. , , which are the populations of molecules with coordination numbers greater than 8 and 11, and the second moment of coordination numbers are derived from the distribution.
Note 3: Distributions of
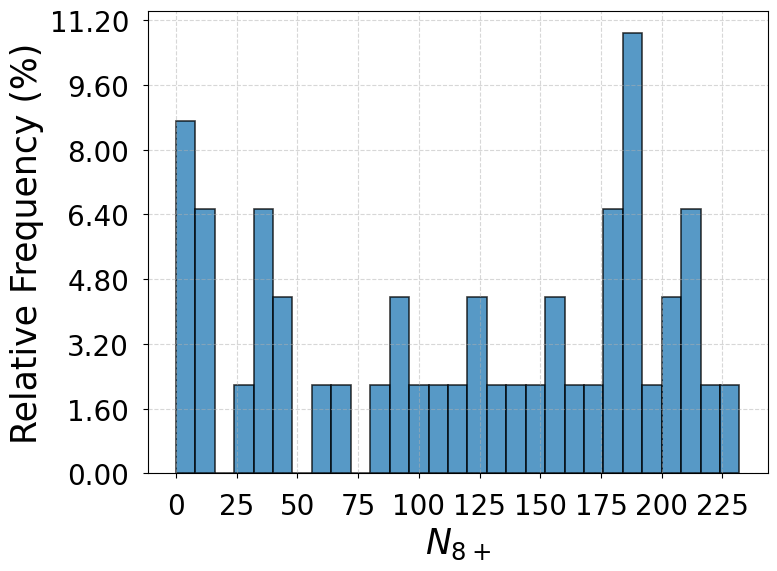
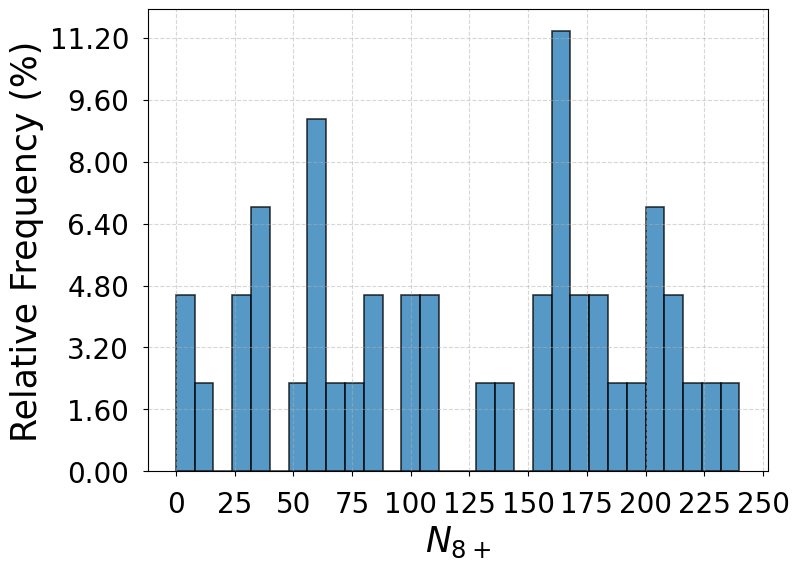
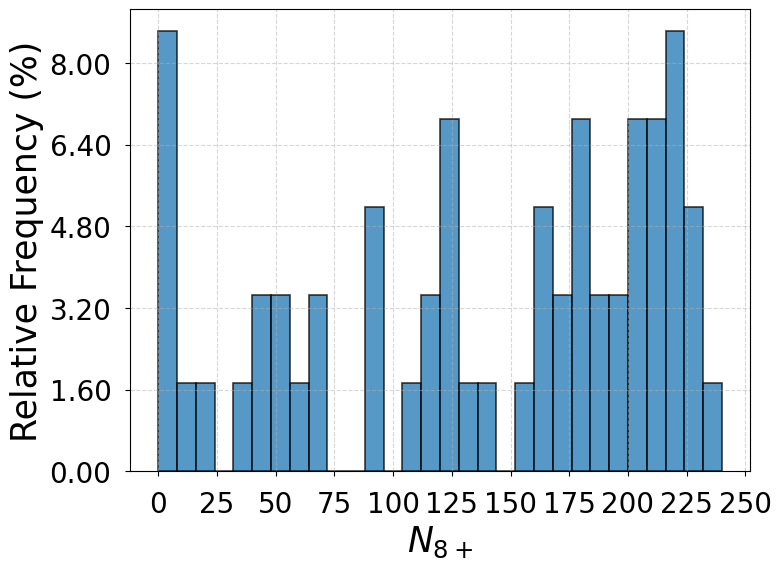
In Supplementary Fig. 1 are the distributions of when the system transitions between the liquid-like basin and the primary crystal basin for the SS model, the GT model, and the full model. Based on the criterion that the liquid-like configurations from WTmetaD simulations count toward the liquid state when their values are within the three-sigma limits of the liquid state distribution drawn from a 50 ns unbiased simulation and otherwise count toward the dense liquid state, the transitions between the liquid-like basin and the primary crystal basin take place directly from or to the liquid state in of the total 46 events for the SS model, of the total 44 events for the GT model, and of the total 58 events for the full model. The other transitions are mediated by the dense liquid state. The results from the three models consistently indicate a predominant two-step nucleation mechanism.
The distributions of beyond the dense liquid state threshold () do not necessarily take similar shapes for the three models. This is because there can exist one or multiple clusters in different configurations and is not accurately proportional to the size of the cluster.