String Sampling with Bidirectional String Anchors
Abstract
The minimizers sampling mechanism is a popular mechanism for string sampling introduced independently by Schleimer et al. [SIGMOD 2003] and by Roberts et al. [Bioinf. 2004]. Given two positive integers and , it selects the lexicographically smallest length- substring in every fragment of consecutive length- substrings (in every sliding window of length ). Minimizers samples are approximately uniform, locally consistent, and computable in linear time. Although they do not have good worst-case guarantees on their size, they are often small in practice. They thus have been successfully employed in several string processing applications. Two main disadvantages of minimizers sampling mechanisms are: first, they also do not have good guarantees on the expected size of their samples for every combination of and ; and, second, indexes that are constructed over their samples do not have good worst-case guarantees for on-line pattern searches.
To alleviate these disadvantages, we introduce bidirectional string anchors (bd-anchors), a new string sampling mechanism. Given a positive integer , our mechanism selects the lexicographically smallest rotation in every length- fragment (in every sliding window of length ). We show that bd-anchors samples are also approximately uniform, locally consistent, and computable in linear time. In addition, our experiments using several datasets demonstrate that the bd-anchors sample sizes decrease proportionally to ; and that these sizes are competitive to or smaller than the minimizers sample sizes using the analogous sampling parameters. We provide theoretical justification for these results by analyzing the expected size of bd-anchors samples. As a negative result, we show that computing a total order on the input alphabet, which minimizes the bd-anchors sample size, is NP-hard.
We also show that by using any bd-anchors sample, we can construct, in near-linear time, an index which requires linear (extra) space in the size of the sample and answers on-line pattern searches in near-optimal time. We further show, using several datasets, that a simple implementation of our index is consistently faster for on-line pattern searches than an analogous implementation of a minimizers-based index [Grabowski and Raniszewski, Softw. Pract. Exp. 2017].
Finally, we highlight the applicability of bd-anchors by developing an efficient and effective heuristic for top- similarity search under edit distance. We show, using synthetic datasets, that our heuristic is more accurate and more than one order of magnitude faster in top- similarity searches than the state-of-the-art tool for the same purpose [Zhang and Zhang, KDD 2020].
1 Introduction
The notion of minimizers, introduced independently by Schleimer et al. [60] and by Roberts et al. [58], is a mechanism to sample a set of positions over an input string. The goal of this sampling mechanism is, given a string of length over an alphabet of size , to simultaneously satisfy the following properties:
- Property 1 (approximately uniform sampling):
-
Every sufficiently long fragment of has a representative position sampled by the mechanism.
- Property 2 (local consistency):
-
Exact matches between sufficiently long fragments of are preserved unconditionally by having the same (relative) representative positions sampled by the mechanism.
In most practical scenarios, sampling the smallest number of positions is desirable, as long as Properties 1 and 2 are satisfied. This is because it leads to small data structures or fewer computations. Indeed, the minimizers sampling mechanism satisfies the property of approximately uniform sampling: given two positive integers and , it selects at least one length- substring in every fragment of consecutive length- substrings (Property 1). Specifically, this is achieved by selecting the starting positions of the smallest length- substrings in every -long fragment, where smallest is defined by a choice of a total order on the universe of length- strings. These positions are called the “minimizers”. Thus from similar fragments, similar length- substrings are sampled (Property 2). In particular, if two strings have a fragment of length in common, then they have at least one minimizer corresponding to the same length- substring. Let us denote by the set of minimizers of string . The following example illustrates the sampling.
Example 1.
The set of minimizers for for string (using a 1-based index) is and for string is . Indeed occurs at position in ; and and have the minimizers and , respectively, which both correspond to string aaa of length .
The minimizers sampling mechanism is very versatile, and it has been employed in various ways in many different applications [47, 67, 19, 10, 31, 35, 36, 48, 37]. Since its inception, the minimizers sampling mechanism has undergone numerous theoretical and practical improvements [56, 10, 54, 53, 15, 22, 72, 37, 74] with a particular focus on minimizing the size of the residual sample; see Section 6 for a summary on this line of research. Although minimizers have been extensively and successfully used, especially in bioinformatics, we observe several inherent problems with setting the parameters and . In particular, although the notion of length- substrings (known as -mers or -grams) is a widely-used string processing tool, we argue that, in the context of minimizers, it may be causing many more problems than it solves: it is not clear to us why one should use an extra sampling parameter to effectively characterize a fragment of length of . In what follows, we describe some problems that may arise when setting the parameters and .
- Indexing:
-
The most widely-used approach is to index the selected minimizers using a hash table. The key is the selected length- substring and the value is the list of positions it occurs. If one would like to use length- substrings for the minimizers with , for some and , they should compute the new set of minimizers and construct their new index based on from scratch.
- Querying:
-
To the best of our knowledge, no index based on minimizers can return in optimal or near-optimal time all occurrences of a pattern of length in .
- Sample Size:
-
If one would like to minimize the number of selected minimizers, they should consider different total orders on the universe of length- strings, which may complicate practical implementations, often scaling only up to a small value, e.g. [22]. On the other hand, when is fixed and increases, the length- substrings in a fragment become increasingly decoupled from each other, and that regardless of the total order we may choose. Unfortunately, this interplay phenomenon is inherent to minimizers. It is known that , for a fixed constant , is a necessary condition for the existence of minimizers samples with expected size in [72]; see Section 6.
We propose the notion of bidirectional string anchors (bd-anchors) to alleviate these disadvantages. The bd-anchors is a mechanism that drops the sampling parameter and its corresponding disadvantages. We only fix a parameter , which can be viewed as the length of the fragments in the minimizers sampling mechanism. The bd-anchor of a string of length is the lexicographically smallest rotation (cyclic shift) of . We unambiguously characterize this rotation by its leftmost starting position in string . The set of the order- bd-anchors of string is the set of bd-anchors of all length- fragments of . It can be readily verified that bd-anchors satisfy Properties 1 and 2.
Example 2.
The set of bd-anchors for for string (using a 1-based index) is and for string , . Indeed occurs at position in ; and and have the bd-anchors and , respectively, which both correspond to the rotation aaaab.
Let us remark that string synchronizing sets, introduced by Kempa and Kociumaka [43], is another string sampling mechanism which may be employed to resolve the disadvantages of minimizers. Yet, it appears to be quite complicated to be efficient in practice. For instance, in [20], the authors used a simplified and specific definition of string synchronizing sets to design a space-efficient data structure for answering longest common extension queries.
We consider the word RAM model of computations with -bit machine words, where , for stating our results. We also assume throughout that string is over alphabet , which captures virtually any real-world scenario. We measure space in terms of -bit machine words. We make the following three specific contributions:
-
1.
In Section 3 we show that the set , for any and any of length , can be constructed in time. We generalize this result showing that for any constant , can be constructed in time using space. Furthermore, we show that the expected size of for strings of length , randomly generated by a memoryless source with identical letter probabilities, is in , for any integer . The latter is in contrast to minimizers which achieve the expected bound of only when , for some constant [72]. We then show, using five real datasets, that indeed the size of decreases proportionally to ; that it is competitive to or smaller than , when ; and that it is much smaller than for small values, which is practically important, as widely-used aligners that are based on minimizers will require less space and computation time if bd-anchors are used instead. Finally, we show a negative result using a reduction from minimum feedback arc set: computing a total order on which minimizes is NP-hard.
-
2.
In Section 4 we show an index based on , for any string of length and any integer , which answers on-line pattern searches in near-optimal time. In particular, for any constant , we show that our index supports the following space/query-time trade-offs:
-
•
it occupies extra space and reports all occurrences of any pattern of length given on-line in time; or
-
•
it occupies extra space and reports all occurrences of any pattern of length given on-line in time.
We also show that our index can be constructed in time. We then show, using five real datasets, that a simple implementation of our index is consistently faster in on-line pattern searches than an analogous implementation of the minimizers-based index proposed by Grabowski and Raniszewski in [31].
-
•
-
3.
In Section 5 we highlight the applicability of bd-anchors by developing an efficient and effective heuristic for top- similarity search under edit distance. This is a fundamental and extensively studied problem [38, 9, 11, 46, 68, 71, 57, 65, 64, 17, 34, 69, 70] with applications in areas including bioinformatics, databases, data mining, and information retrieval. We show, using synthetic datasets, that our heuristic, which is based on the bd-anchors index, is more accurate and more than one order of magnitude faster in top- similarity searches than the state-of-the-art tool proposed by Zhang and Zhang in [70].
In Section 2, we provide some preliminaries; and in Section 6 we discuss works related to minimizers. Let us stress that, although other works may be related to our contributions, we focus on comparing to minimizers because they are extensively used in applications. The source code of our implementations is available at https://github.com/solonas13/bd-anchors.
A preliminary version of this paper appeared as [49].
2 Preliminaries
We start with some basic definitions and notation following [13]. An alphabet is a finite nonempty set of elements called letters. A string is a sequence of length of letters from . The empty string, denoted by , is the string of length . The fragment of is an occurrence of the underlying substring . We also say that occurs at position in . A prefix of is a fragment of of the form and a suffix of is a fragment of of the form . The set of all strings over (including ) is denoted by . The set of all length- strings over is denoted by . Given two strings and , the edit distance is the minimum number of edit operations (letter insertion, deletion, or substitution) transforming one string into the other.
Let be a finite nonempty set of strings over of total length . We define the trie of , denoted by , as a deterministic finite automaton that recognizes . Its set of states (nodes) is the set of prefixes of the elements of ; the initial state (root node) is ; the set of terminal states (leaf nodes) is ; and edges are of the form , where and are nodes and . The size of is thus . The compacted trie of , denoted by , contains the root node, the branching nodes, and the leaf nodes of . The term compacted refers to the fact that reduces the number of nodes by replacing each maximal branchless path segment with a single edge, and that it uses a fragment of a string to represent the label of this edge in machine words. The size of is thus . When is the set of suffixes of a string , then is called the suffix tree of , and we denote it by . The suffix tree of a string of length over an alphabet can be constructed in time [23].
Let us fix throughout a string of length over an ordered alphabet . Recall that we make the standard assumption of an integer alphabet .
We start by defining the notion of minimizers of from [58] (the definition in [60] is slightly different). Given an integer , an integer , and the th length- fragment of , we define the -minimizers of as the positions where a lexicographically minimal length- substring of occurs. The set of -minimizers of is defined as the set of -minimizers of , for all . The density of is defined as the quantity . The following bounds are obtained trivially. The density of any minimizer scheme is at least , since at least one -minimizer is selected in each fragment, and at most , when every -minimizer is selected.
If we waive the lexicographic order assumption, the set can be computed on-line in time, and if we further assume a constant-time computable function that gives us an arbitrary rank for each length- substring in in constant amortized time [37]. This can be implemented, for instance, using a rolling hash function (e.g. Karp-Rabin fingerprints [41]), and the rank (total order) is defined by this function. We also provide here, for completeness, a simple off-line -time algorithm that uses a lexicographic order.
Theorem 1.
The set , for any integers and any string of length , can be constructed in time.
Proof.
The underlying algorithm has two main steps. In the first step, we construct , the suffix tree of in time [23]. Using a depth-first search traversal of we assign at every position of in the lexicographic rank of among all the length- strings occurring in . This process clearly takes time as is an ordered structure; it yields an array of size with lexicographic ranks. In the second step, we apply a folklore algorithm, which computes the minimum elements in a sliding window of size (cf. [37]) over . The set of reported indices is . ∎
3 Bidirectional String Anchors
We introduce the notion of bidirectional string anchors (bd-anchors). Given a string , a string is a rotation (or cyclic shift or conjugate) of if and only if there exists a decomposition such that , for a string and a nonempty string . We often characterize by its starting position in . We use the term rotation interchangeably to refer to string or to its identifier .
Definition 1 (Bidirectional anchor).
Given a string of length , the bidirectional anchor (bd-anchor) of is the lexicographically minimal rotation of with minimal . The set of order- bd-anchors of a string of length , for some integer , is defined as the set of bd-anchors of , for all .
The density of is defined as the quantity . It can be readily verified that the bd-anchors sampling mechanism satisfies Properties 1 (approximately uniform sampling) and 2 (local consistency).
Example 3.
Let , , and . Strings and (are at Hamming distance 2 but) have the same set of bd-anchors of order : . The reader can probably share the intuition that the bd-anchors sampling mechanism is suitable for sequence comparison due to Properties 1 and 2, in particular, when the parameter is set accordingly.
Linear-Time Construction of .
Importantly, we show that admits an efficient construction. One can use the linear-time algorithm by Booth [6] to compute the lexicographically minimal rotation for each length- fragment of , resulting in an -time algorithm, which is reasonably fast for modest . (Booth’s algorithm gives the leftmost minimal rotation by construction.) We instead give an optimal -time algorithm for the construction of , which is mostly of theoretical interest.
For every string and every natural number , we define the th power of the string , denoted by , by and for . A nonempty string is primitive, if it is not the power of any other string. Let us state two well-known combinatorial lemmas.
Lemma 1 ([13]).
A nonempty string is primitive if and only if it occurs as a substring in only as a prefix and as a suffix.
Lemma 2 ([61]).
Let and , for two strings . If is primitive then is also primitive.
A substring of a string is called an infix of if and only if with and .
Lemma 3.
A string has more than one minimal lexicographic rotation if and only if is a power of some string.
Proof.
-
Let , and be the leftmost minimal lexicographic rotation of . Suppose towards a contradiction that has another minimal lexicographic rotation but is primitive. In particular, there exists , with and . If is primitive, then is also primitive by Lemma 2 but then has occurring as infix. In particular, in , is a suffix of the first occurrence of and is a prefix of and thus occurs as infix. By Lemma 1 we obtain a contradiction.
-
Let and a minimal lexicographic rotation of be . Then either or is a minimal lexicographic rotation of .
∎
Example 4 (Illustration of Lemma 3).
Let , with and , and with and . Observe that occurs as infix (shown as underlined) in hence is a power of some string.
Lemma 4.
Let be a string of length and set , for some letter not occurring in that is the lexicographically maximal letter occurring in . Further let be the lexicographically minimal suffix of , for some . The leftmost lexicographically minimal rotation of is .
Proof.
First note that because is the lexicographically maximal letter occurring in .
We consider two cases: (i) is primitive; and (ii) is power of some string. In the first case, has one lexicographically minimal rotation by Lemma 3, and thus this is . In the second case, has more than one lexicographically minimal rotations, but because is power of some string and is the lexicographically maximal letter occurring in , is the leftmost lexicographically minimal rotation of . ∎
We employ the data structure of Kociumaka [44, Theorem 20] to obtain the following result.
Theorem 2.
The set , for any and any of length , can be constructed in time.
Proof.
The data structure of Kociumaka [44, Theorem 20] gives the minimal lexicographic suffix for any concatenation of arbitrary fragments of a string in time after an time preprocessing.
We set , for some letter that does not occur in and is the lexicographically maximal letter occurring in . For each fragment , we compute the minimal lexicographic suffix of string
where in time. This suffix of is the minimal lexicographic rotation by Lemma 4. ∎
Space-Efficient Construction of .
It should be clear that, in the best case, the size of is in and this bound is tight. The construction of Theorem 2 requires space. Ideally, we would thus like to compute efficiently using (strongly) sublinear space. We generalize Theorem 2 to the following result.
Theorem 3.
The set , for any , any of length , and any constant , can be constructed in time using space.
Proof.
We compute for all using the algorithm from Theorem 2 and output their union. For any constant , the alphabet size is still polynomial in the length of the fragments, so computing one such anchor set takes time and space by Theorem 2. We delete each fragment (and the associated data structure) before processing the subsequent anchor set: it takes time and additional space to construct . ∎
Expected Size of .
We next analyze the expected size of . We first show that if grows no faster than the size of the alphabet, then the expected size of is in . Otherwise, if grows faster than , we slightly amend the sampling process to ensure that the expected size of the sample is in .
Lemma 5.
If is a string of length , randomly generated by a memoryless source over an alphabet of size with identical letter probabilities, then, for any integer , the expected size of is in .
Proof.
If , then . If , then . Now suppose . We say that introduces a new bd-anchor if there exists such that is the bd-anchor of , but is not the bd-anchor of for all . Let denote the event that introduces a new bd-anchor. Since the letters are independent identically distributed, the probability only depends on and is non-increasing in the number of preceding overlapping length- substrings. Therefore
Let be the length of the shortest prefix of the lexicographically minimal rotation of which is strictly smaller than the same length prefix of any other rotation of .
Note that
| (4) | |||||
To bound the probability in (4), note that
The probability in (4) is bounded by since each letter of a primitive length- string is equally likely to be the anchor. Similarly, the probability in (4) is bounded by . Finally, the probability in (4) is bounded by the probability that two prefixes of length of rotations of are equal, which is at most . It follows that
We conclude that for any the expected size of is in . ∎
We define a reduced version of bd-anchors to ensure that the expected size of the sample is in .
Definition 2.
Given a string of length and an integer , we define the reduced bidirectional anchor of as the lexicographically minimal rotation of with minimal . The set of order- reduced bd-anchors of a string of length is defined as the set of reduced bd-anchors of , for all .
Lemma 6.
If is a string of length , randomly generated by a memoryless source over an alphabet of size with identical letter probabilities, then, for any integer , the expected size of with is in .
Proof.
If , then . Now suppose . Analogously to in 5, we denote the event that introduces a new reduced bd-anchor by . Again we find
Let be the length of the shortest prefix of the lexicographically minimal rotation of which is strictly smaller than the same length prefix of any other rotation . Using a similar proof to that of 5 we find that
We conclude that for any the expected size of is in . ∎
In particular, if , we employ the sampling mechanism underlying , otherwise () we employ the sampling mechanism underlying with to ensure that the expected density of the residual sampling is always in .
Density Evaluation.
We compare the density of bd-anchors and reduced bd-anchors, denoted by BDA and rBDA, respectively, to the density of minimizers, for different values of and such that . This is a fair comparison because is the length of the fragments considered by both mechanisms. We implemented bd-anchors, the standard minimizers mechanism from [58], and the minimizers mechanism with robust winnowing from [60]. The standard minimizers and those with robust winnowing are referred to as STD and WIN, respectively.
For bd-anchors, we used Booth’s algorithm, which is easy to implement and reasonably fast. For minimizers, we used Karp-Rabin fingerprints [41]. (Note that such “random” minimizers tend to perform even better than the ones based on lexicographic total order in terms of density [72].) For the reduced version of bd-anchors, we used , because the value suggested by Lemma 6 is relatively large for the small values tested; e.g. for , . Throughout, we do not evaluate construction times, as all implementations are reasonably fast, and we make the standard assumption that preprocessing is only required once. We used five string datasets from the popular Pizza & Chili corpus [25] (see Table 1 for the datasets characteristics). All implementations referred to in this paper have been written in C++ and compiled at optimization level -O3. All experiments reported in this paper were conducted using a single core of an AMD Opteron 6386 SE 2.8GHz CPU and 252GB RAM running GNU/Linux.
| Dataset | Length | Alphabet |
|---|---|---|
| size | ||
| DNA | 200,000,000 | 4 |
| XML | 200,000,000 | 95 |
| ENGLISH | 200,000,000 | 224 |
| PROTEINS | 200,000,000 | 27 |
| SOURCES | 200,000,000 | 229 |





As can be seen by the results depicted in Figures 1 and 2, the density of both BDA and rBDA is either significantly smaller than or competitive to the STD and WIN minimizers density, especially for small . This is useful because a lower density results in smaller indexes and less computation (see Section 4), and because small is of practical interest (see Section 5). For instance, the widely-used long-read aligner Minimap2 [48] stores the selected minimizers of a reference genome in a hash table to find exact matches as anchors for seed-and-extend alignment. The parameters and are set based on the required sensitivity of the alignment, and thus and cannot be too large for high sensitivity. Thus, a lower sampling density reduces the size of the hash table, as well as the computation time, by lowering the average number of selected minimizers to consider when performing an alignment. Furthermore, although the datasets are not uniformly random, rBDA performs better than BDA as grows, as suggested by Lemmas 5 and 6.
There exists a long line of research on improving the density of minimizers in special regimes (see Section 6 for details). We stress that most of these algorithms are designed, implemented, or optimized, only for the DNA alphabet. We have tested against two state-of-the-art tools employing such algorithms: Miniception [72] and PASHA [22]. The former did not give better results than STD or WIN for the tested values of and ; and the latter does not scale beyond or with large alphabets. We have thus omitted these results.
We next report the average number (AVG) of bd-anchors of order over all strings of length (see Table 2(a)) and over all strings of length (see Table 2(b)), both over a binary alphabet. Notably, the results show that AVG always lies in even if not using the reduced version of bd-anchors (see Lemma 6). As expected by Lemma 5, the analogous AVG values using a ternary alphabet (not reported here) were always lower than the corresponding ones with a binary alphabet.
| 5 | 2.5 | 1.66 | 1.25 | |
| AVG | 8.53 | 4.37 | 2.77 | 1.76 |
| 16 | 8 | 5.33 | 4 |
| 8 | 4 | 2,66 | 2 | |
| AVG | 14.16 | 7.67 | 5.26 | 3.85 |
| 16 | 8 | 5.33 | 4 |
Minimizing the Size of is NP-hard.
The number of bd-anchors depends on the lexicographic total order defined on . We now prove that finding the total order which minimizes the number of bd-anchors is NP-hard using a reduction from minimum feedback arc set [40]. Let us start be defining this problem. Given a directed graph , a feedback arc set in is a subset of that contains at least one edge from every cycle in . Removing these edges from breaks all of the cycles, producing a directed acyclic graph. In the minimum feedback arc set problem, we are given , and we are asked to compute a smallest feedback arc set in . The decision version of the problem takes an additional parameter as input, and it asks whether all cycles can be broken by removing at most edges from . The decision version is NP-complete [40] and the optimization version is APX-hard [21].
Theorem 4.
Let be a string of length over alphabet . Further let be an integer. Computing a total order on which minimizes is NP-hard.
Proof.
Let be any instance of the minimum feedback arc set problem. We will construct a string in polynomial time in the size of such that finding a total order on which minimizes corresponds to finding a minimum feedback arc set in .
We start with an empty string . For each edge , we append to . Observe that, if , all the ’s of and none of its ’s are order-4 bd-anchors, except possibly the first and last depending on the preceding and subsequent letters in . Thus there are to order- bd-anchors in . If on the other hand , we analogously find that there will be to order- bd-anchors in .
Let be the number of edges such that . The total number of order- bd-anchors in is
with . Therefore minimizing the total number of order- bd-anchors in is equivalent to finding an order on the set of vertices of which minimizes .
Note that if we delete all edges such that , then the residual graph is acyclic. Moreover, for each acyclic graph there exists an order on the vertices such that for all . Therefore the minimal equals the size of the minimum feedback arc set.
We conclude that, since finding the size of the minimum feedback arc set is NP-hard, so is finding a total order on which minimizes the total number of order-4 bd-anchors. ∎
4 Indexing Using Bidirectional Anchors
Before presenting our index, let us start with a basic definition that is central to our querying process.
Definition 3 (-hit).
Given an order- bd-anchor , for some integer , of a query string , two integers , with , and an order- bd-anchor of a target string , the ordered pair is called an -hit if and only if and .
Intuitively, the parameters and let us choose a fragment of that is anchored at .
Example 5.
Let , , and . Consider that we would like to find the common fragment . We know that the bd-anchor of order corresponding to is , and thus to find it we set and . The ordered pair is a -hit because for , we have: and .
We would like to construct a data structure over , which is based on , such that, when we are given an order- bd-anchor over as an on-line query, together with parameters and , we can report all -hits efficiently. To this end, we present an efficient data structure, denoted by , which is constructed on top of , and answers -hit queries in near-optimal time. We prove the following result.
Theorem 5.
Given a string of length and an integer , the index can be constructed in time. For any constant , :
-
•
occupies extra space and reports all -hits in time; or
-
•
occupies extra space and reports all -hits in time.
Let us denote by the reversal of string . We now describe our data structure.
Construction of .
Given , we construct two sets and of strings; conceptually, the reversed suffixes going left from to , and the suffixes going right from to , for all in . In particular, for the bd-anchor , we construct two strings: and . Note that, , since for every bd-anchor in we have a distinct string in and in .
We construct two compacted tries and over and , respectively, to index all strings. Every string is concatenated with some special letter not occurring in , which is lexicographically minimal, to make and prefix-free (this is standard for conceptual convenience). The leaf nodes of the compacted tries are labeled with the corresponding : there is a one-to-one correspondence between a leaf node and a bd-anchor . In time, we also enhance the nodes of the tries with a perfect static dictionary [27] to ensure constant-time retrieval of edges by the first letter of their label. Let denote the list of the leaf labels of as they are visited using a depth-first search traversal. corresponds to the (labels of the) lexicographically sorted list of in increasing order. For each node in , we also store the corresponding interval over . Analogously for , denotes the list of the leaf labels of as they are visited using a depth-first search traversal and corresponds to the (labels of the) lexicographically sorted list of in increasing order. For each node in , we also store the corresponding interval over .
The total size occupied by the tries is because they are compacted: we label the edges with intervals over from .
We also construct a 2D range reporting data structure over the following points in set :
Note that because the set of leaf labels stored in both tries is precisely the set . Let us remark that the idea of employing 2D range reporting for bidirectional pattern searches has been introduced by Amir et al. [2] for text indexing and dictionary matching with one error; see also [50].
This completes the construction of . We next explain how we can query .
Querying.
Given a bd-anchor over a string as an on-line query and parameters , we spell in and in starting from the root nodes. If any of the two strings is not spelled fully, we return no -hits. If both strings are fully spelled, we arrive at node in (resp. in ), which corresponds to an interval over stored in (resp. in ). We obtain the two intervals and forming a rectangle and ask the corresponding 2D range reporting query. It can be readily verified that this query returns all -hits.
Example 6.
Let and . We have the following strings in : ; ; ; and . We have the following strings in : ; ; ; . Inspect Figure 3.


Proof of Theorem 5.
We use the -time algorithm underlying Theorem 2 to construct . We use the -time algorithm from [3, 8] to construct the compacted tries from . We extract the points using the compacted tries in time. For the first trade-off of the statement, we use the -time algorithm from [5] to construct the 2D range reporting data structure over from [7]. For the second trade-off, we use the -time algorithm from [29] to construct the 2D range reporting data structure over from the same paper. ∎
We obtain the following corollary for the fundamental problem of text indexing [66, 51, 23, 39, 24, 32, 33, 4, 12, 55, 43, 28].
Corollary 6.
Given constructed for some integer and some constant over string , we can report all occurrences of any pattern , , in in time:
-
•
when occupies extra space; or
-
•
when occupies extra space.
Proof.
Every occurrence of in is prefixed by string . We first compute the bd-anchor of in time using Booth’s algorithm. Let this bd-anchor be . We set and . The result follows by applying Theorem 5. ∎
Querying Multiple Fragments.
In the case of approximate pattern matching, we may want to query multiple length- fragments of a string given as an on-line query, and not only its length- prefix. We show that such an operation can be done efficiently using the bd-anchors of and the index.
Corollary 7.
Given constructed for some and some constant over , for any sequence (not necessarily consecutive) of length- fragments of a pattern , , corresponding to the same order- bd-anchor of , we can report all occurrences of all fragments in in time:
-
•
when occupies space; or
-
•
when occupies space.
Proof.
Let the order- bd-anchor over be and the corresponding parameters be , with . Observe that and . Starting from , the string we spell for fragment is the prefix of for fragment . The analogous property holds for the other direction: the string we spell for fragment is the prefix of for fragment . Thus it takes only time to construct all rectangles. Finally, we ask the corresponding 2D range reporting queries to obtain all occurrences in the claimed time complexities. ∎
Index Evaluation.
Consider a hash table with the following (key, value) pairs: the key is the hash value of a length- string ; and the value (satellite data) is a list of occurrences of in . It should be clear that such a hash table indexing the minimizers of does not perform well for on-line pattern searches of arbitrary length because it would need to verify the remaining prefix and suffix of the pattern using letter comparisons for all occurrences of a minimizer in . We thus opted for comparing our index to the one of [31], which addresses this specific problem by sampling the suffix array [51] with minimizers to reduce the number of letter comparisons during verification.
To ensure a fair comparison, we have implemented the basic index from [31]; we denote it by GR Index. We used Karp-Rabin [41] fingerprints for computing the minimizers of . We also used the array-based version of the suffix tree that consists of the suffix array (SA) and the longest common prefix (LCP) array [51]; SA was constructed using SDSL [30] and the LCP array using the Kasai et al. [42] algorithm.
We sampled the SA using the minimizers. Given a pattern , we searched starting with the minimizer using the Manber and Myers [51] algorithm on the sampled SA. For verifying the remaining prefix of , we used letter comparisons, as described in [31]. The space complexity of this implementation is and the extra space for the index is . The query time is not bounded. We have implemented two versions of our index. We used Booth’s algorithm for computing the bd-anchors of . We used SDSL for SA construction and the Kasai et al. algorithm for LCP array construction. We sampled the SA using the bd-anchors thus constructing and . Then, the two versions of our index are:
-
1.
BDA Index v1: Let be the bd-anchor of . For (resp. ) we used the Manber and Myers algorithm for searching over (resp. ). We used range trees [14] implemented in CGAL [63] for 2D range reporting as per the described querying process. The space complexity of this implementation is and the extra space for the index is . The query time is , where is the total number of occurrences of in .
-
2.
BDA Index v2: Let be the bd-anchor of . If (resp. ), we search for (resp. ) using the Manber and Myers algorithm on (resp. ). For verifying the remaining part of the pattern we used letter comparisons. The space complexity of this implementation is and the extra space for the index is . The query time is not bounded.
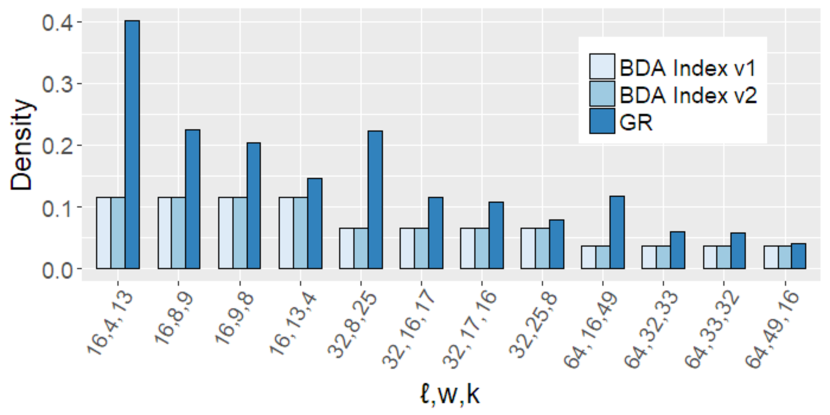
For each of the five real datasets of Table 1 and each query string length , we randomly extracted 500,000 substrings from the text and treated each substring as a query, following [31]. We plot the average query time in Figure 4. As can be seen, BDA Index v2 consistently outperforms GR Index across all datasets and all values. The better performance of BDA Index v2 is due to two theoretical reasons. First, the verification strategy exploits the fact that the index is bidirectional to apply the Manber and Myers algorithm to the largest part of the pattern, which results in fewer letter comparisons. Second, bd-anchors generally have smaller density compared to minimizers; see Figure 5. We also plot the peak memory usage in Figure 6. As can be seen, BDA Index v2 requires a similar amount of memory to GR Index.





BDA Index v1 was slower than GR Index for small but faster for large in three out of five datasets used and had by far the highest memory usage. Let us stress that the inefficiency of BDA Index v1 is not due to inefficiency in the query time or space of our algorithm. It is merely because the range tree implementation of CGAL, which is a standard off-the-shelf library, is unfortunately inefficient in terms of both query time and memory usage; see also [62, 26].









Discussion.
The proposed index, which is based on bd-anchors, has the following attributes:
-
1.
Construction: is constructed in worst-case time and is constructed in worst-case time. These time complexities are near-linear in and do not depend on the alphabet as long as , which is true for virtually any real scenario.
- 2.
-
3.
Querying: The index answers on-line pattern searches in near-optimal time.
-
4.
Flexibility: Note that one would have to reconstruct a (hash-based) index, which indexes the set of -minimizers, to increase specificity or sensitivity: increasing increases the specificity and decreases the sensitivity. Our index, conceptually truncated at string depth , is essentially an index based on -minimizers, which additionally wrap around. We can thus increase specificity by considering larger values or increase sensitivity by considering smaller values. This effect can be realized without reconstructing our index: we just adapt and upon querying accordingly.
5 Similarity Search under Edit Distance
We show how bd-anchors can be applied to speed up similarity search under edit distance. This is a fundamental problem with myriad applications in bioinformatics, databases, data mining, and information retrieval. It has thus been studied extensively in the literature both from a theoretical and a practical point of view [38, 9, 11, 46, 68, 71, 57, 65, 64, 17, 34, 69, 70]. Let be a collection of strings called dictionary. We focus, in particular, on indexing for answering the following type of top- queries: Given a query string and an integer , return strings from the dictionary that are closest to with respect to edit distance. We follow a typical seed-chain-align approach as used by several bioinformatics applications [1, 16, 47, 48]. The main new ingredients we inject, with respect to this classic approach, is that we use: (1) bd-anchors as seeds; and (2) to index the dictionary , for some integer parameter .
Construction.
We require an integer parameter defining the order of the bd-anchors. We set , where , compute the bd-anchors of order of , and construct the index (see Section 4) using the bd-anchors.
Querying.
We require two parameters and . The former parameter controls the sensitivity of our filtering step (Step 2 below); and the latter one controls the sensitivity of our verification step (Step 3 below). Both parameters trade accuracy for speed.
-
1.
For each query string , we compute the bd-anchors of order . For every bd-anchor , we take an arbitrary fragment (e.g. the leftmost) of length anchored at as the seed. Let this fragment start at position . This implies a value for and , with ; specifically for we have and . For every bd-anchor , we query in and in and collect all -hits.
-
2.
Let be an input parameter and let be the list of all -hits between the queried fragments of string and fragments of a string . If , we consider string as not found. The intuition here is that if and are sufficiently close with respect to edit distance, they would have a relatively long [16]. If , we sort the elements of with respect to their first component. (This comes for free because we process from left to right.) We then compute a longest increasing subsequence (LIS) in with respect to the second component, which chains the -hits, in time [59] per list. We use the LIS of to estimate the identity score (total number of matching letters in a fixed alignment) for and , which we denote by , based on the occurrences of the -hits in the LIS.
-
3.
Let be an input parameter and let be the th largest estimated identity score. We extract, as candidates, the ones whose estimated identity score is at least . For every candidate string , we close the gaps between the occurrences of the -hits in the LIS using dynamic programming [45], thus computing an upper bound on the edit distance between and (UB score). In particular, closing the gaps consists in summing up the exact edit distance for all pairs of fragments (one from and one from ) that lie in between the -hits. We return strings from the list of candidates with the lowest UB score. If , we return strings with the highest score.
Index Evaluation.
We compared our algorithm, called BDA Search, to Min Search, the state-of-the-art tool for top- similarity search under edit distance proposed by Zhang and Zhang in [70]. The main concept used in Min Search is the rank of a letter in a string, defined as the size of the neighborhood of the string in which the letter has the minimum hash value. Based on this concept, Min Search partitions each string in the dictionary into a hierarchy of substrings and then builds an index comprised of a set of hash tables, so that strings having common substrings and thus small edit distance are grouped into the same hash table. To find the top- closest strings to a query string, Min Search partitions the query string based on the ranks of its letters and then traverses the hash tables comprising the index. Thanks to the index and the use of several filtering tricks, Min Search is at least one order of magnitude faster with respect to query time than popular alternatives [69, 71, 18].
We implemented two versions of BDA Search: BDA Search v1 which is based on BDA Index v1; and BDA Search v2 which is based on BDA Index v2. For Min Search, we used the C++ implementation from https://github.com/kedayuge/Search.
We constructed synthetic datasets, referred to as SYN, in a way that enables us to study the impact of different parameters and efficiently identify the ground truth (top- closest strings to a query string with respect to edit distance). Specifically, we first generated query strings and then constructed a cluster of strings around each query string. To generate the query strings, we started from an arbitrary string of length from a real dataset of protein sequences, used in [70], and generated a string that is at edit distance from , by performing edit distance operations, each with equal probability. Then, we treated as and repeated the process to generate the next query string. To create the clusters, we first added each query string into an initially empty cluster and then added strings, each at edit distance at most from the query string. The strings were generated by performing at most edit distance operations, each with equal probability. Thus, each cluster contains the top- closest strings to the query string of the cluster. We used , , and . We evaluated query answering accuracy using the F1 score [52], expressed as the harmonic mean of precision and recall111Precision is the ratio between the number of returned strings that are among the top- closest strings to a query string and the number of all returned strings. Recall is the ratio between the number of returned strings that are among the top- closest strings to a query string and . Since all tested algorithms return strings, the F1 score in our experiments is equal to precision and equal to recall.. For BDA Search, we report results for (full sensitivity during filtering) and (no sensitivity during verification), as it was empirically determined to be a reasonable trade-off between accuracy and speed. For Min Search, we report results using its default parameters from [70].
We plot the F1 scores and average query time in Figures 7 and 8, respectively. All methods achieved almost perfect accuracy, in all tested cases. BDA Search slightly outperformed Min Search (by up to ), remaining accurate even for large ; the changes to F1 score for Min Search as varies are because the underlying method is randomized. However, both versions of BDA Search were more than one order of magnitude faster than Min Search on average (over all results of Figure 8), with BDA Search v1 being 2.9 times slower than BDA Search v2 on average, due to the inefficiency of the range tree implementation of CGAL. Furthermore, both versions of BDA Search scaled better with respect to . For example, the average query time for BDA Search v1 became 2 times larger when increased from to (on average over values), while that for Min Search became 5.4 times larger on average. The reason is that verification in Min Search, which increases the accuracy of this method, becomes increasingly expensive as gets larger. The peak memory usage for these experiments is reported in Figure 9. Although Min Search outperforms BDA Search in terms of memory usage, BDA Search v2 still required a very small amount of memory (less than 1GB). BDA Search v1 required more memory for the reasons mentioned in Section 4.






Discussion.
BDA Search outperforms Min Search in accuracy while being more than one order of magnitude faster in query time. These results are very encouraging because the efficiency of BDA Search is entirely due to injecting bd-anchors and not due to any further filtering tricks such as those employed by Min Search. Min Search clearly outperforms BDA Search in memory usage, albeit the memory usage of BDA Search v2 is still quite modest. We defer an experimental evaluation using real datasets to the journal version of our work.
6 Other Works on Improving Minimizers
Although every sampling mechanism based on minimizers primarily aims at satisfying Properties 1 and 2, different mechanisms employ total orders that lead to substantially different total numbers of selected minimizers. Thus, research on minimizers has focused on determining total orders which lead to the lowest possible density (recall that the density is defined as the number of selected length- substrings over the length of the input string). In fact, much of the literature focuses on the average case [56, 54, 53, 22, 72]; namely, the lowest expected density when the input string is random. In practice, many works use a “random minimizer” where the order is defined by choosing a permutation of all the length- strings at random (e.g. by using a hash function, such as the Karp-Rabin fingerprints [41], on the length- strings). Such a randomized mechanism has the benefit of being easy to implement and providing good expected performance in practice.
Minimizers and Universal Hitting Sets.
A universal hitting set (UHS) is an unavoidable set of length- strings, i.e., it is a set of length- strings that “hits” every -long fragment of every possible string. The theory of universal hitting sets [56, 53, 43, 73] plays an important role in the current theory for minimizers with low density on average. In particular, if a UHS has small size, it generates minimizers with a provable upper-bound on their density. However, UHSs are less useful in the string-specific case for two reasons [74]: (1) the requirement that a UHS has to hit every -long fragment of every possible string is too strong; and (2) UHSs are too large to provide a meaningful upper-bound on the density in the string-specific case. Therefore, since in many practical scenarios the input string is known and does not change frequently, we try to optimize the density for one particular string instead of optimizing the average density over a random input.
String-Specific Minimizers.
In the string-specific case, minimizers sampling mechanisms may employ frequency-based orders [10, 37]. In these orders, length- strings occurring less frequently in the string compare less than the ones occurring more frequently. The intuition [74] is to obtain a sparse sampling by selecting infrequent length- strings which should be spread apart in the string. However, there is no theoretical guarantee that a frequency-based order gives low density minimizers (there are many counter-examples). Furthermore, frequency-based orders do not always give minimizers with lower density in practice. For instance, the two-tier classification (very frequent vs. less frequent length- strings) in the work of [37] outperforms an order that strictly follows frequency of occurrence.
A different approach to constructing string-specific minimizers is to start from a UHS and to remove elements from it, as long as it still hits every -long fragment of the input string [15]. Since this approach starts with a UHS that is not related to the string, the improvement in density may not be significant [74]. Additionally, current methods [22] employing this approach are computationally limited to using , as the size of the UHS increases exponentially with . Using such small values may not be appropriate in some applications.
Other Improvements.
When , minimizers with expected density of on a random string can be constructed using the approach of [72]. Such minimizers have guaranteed expected density less than and work for infinitely many and . The approach of [72] also does not require the use of expensive heuristics to precompute and store a large set of length- strings, unlike some methods [56, 15, 22] with low density in practice.
The notion of polar set, which can be seen as complementary to that of UHS, was recently introduced in [74]. While a UHS is a set of length- strings that intersect with every -long fragment at least once, a polar set is a set of length- strings that intersect with any fragment at most once. The construction of a polar set builds upon sets of length- strings that are sparse in the input string. Thus, the minimizers derived from these polar sets have provably tight bounds on their density. Unfortunately, computing optimal polar sets is NP-hard, as shown in [74]. Thus, the work of [74] also proposed a heuristic for computing feasible “good enough” polar sets. A main disadvantage of this approach is that when each length- string occurs frequently in the input string, it becomes hard to select many length- strings without violating the polar set condition.
References
- [1] Stephen F. Altschul, Warren Gish, Webb Miller, Eugene W. Myers, and David J. Lipman. Basic local alignment search tool. Journal of Molecular Biology, 215(3):403–410, 1990. doi:10.1016/S0022-2836(05)80360-2.
- [2] Amihood Amir, Dmitry Keselman, Gad M. Landau, Moshe Lewenstein, Noa Lewenstein, and Michael Rodeh. Text indexing and dictionary matching with one error. J. Algorithms, 37(2):309–325, 2000. URL: https://doi.org/10.1006/jagm.2000.1104, doi:10.1006/jagm.2000.1104.
- [3] Carl Barton, Tomasz Kociumaka, Chang Liu, Solon P. Pissis, and Jakub Radoszewski. Indexing weighted sequences: Neat and efficient. Inf. Comput., 270, 2020. URL: https://doi.org/10.1016/j.ic.2019.104462, doi:10.1016/j.ic.2019.104462.
- [4] Djamal Belazzougui. Linear time construction of compressed text indices in compact space. In Symposium on Theory of Computing, STOC 2014, New York, NY, USA, May 31 - June 03, 2014, pages 148–193, 2014. doi:10.1145/2591796.2591885.
- [5] Djamal Belazzougui and Simon J. Puglisi. Range predecessor and lempel-ziv parsing. In Robert Krauthgamer, editor, Proceedings of the Twenty-Seventh Annual ACM-SIAM Symposium on Discrete Algorithms, SODA 2016, Arlington, VA, USA, January 10-12, 2016, pages 2053–2071. SIAM, 2016. URL: https://doi.org/10.1137/1.9781611974331.ch143, doi:10.1137/1.9781611974331.ch143.
- [6] Kellogg S. Booth. Lexicographically least circular substrings. Inf. Process. Lett., 10(4/5):240–242, 1980. URL: https://doi.org/10.1016/0020-0190(80)90149-0, doi:10.1016/0020-0190(80)90149-0.
- [7] Timothy M. Chan, Kasper Green Larsen, and Mihai Patrascu. Orthogonal range searching on the RAM, revisited. In Ferran Hurtado and Marc J. van Kreveld, editors, Proceedings of the 27th ACM Symposium on Computational Geometry, Paris, France, June 13-15, 2011, pages 1–10. ACM, 2011. URL: https://doi.org/10.1145/1998196.1998198, doi:10.1145/1998196.1998198.
- [8] Panagiotis Charalampopoulos, Costas S. Iliopoulos, Chang Liu, and Solon P. Pissis. Property suffix array with applications in indexing weighted sequences. ACM J. Exp. Algorithmics, 25, April 2020. URL: https://doi.org/10.1145/3385898, doi:10.1145/3385898.
- [9] Surajit Chaudhuri, Kris Ganjam, Venkatesh Ganti, and Rajeev Motwani. Robust and efficient fuzzy match for online data cleaning. In Alon Y. Halevy, Zachary G. Ives, and AnHai Doan, editors, Proceedings of the 2003 ACM SIGMOD International Conference on Management of Data, San Diego, California, USA, June 9-12, 2003, pages 313–324. ACM, 2003. URL: https://doi.org/10.1145/872757.872796, doi:10.1145/872757.872796.
- [10] Rayan Chikhi, Antoine Limasset, and Paul Medvedev. Compacting de Bruijn graphs from sequencing data quickly and in low memory. Bioinform., 32(12):201–208, 2016. URL: https://doi.org/10.1093/bioinformatics/btw279, doi:10.1093/bioinformatics/btw279.
- [11] Richard Cole, Lee-Ad Gottlieb, and Moshe Lewenstein. Dictionary matching and indexing with errors and don’t cares. In László Babai, editor, Proceedings of the 36th Annual ACM Symposium on Theory of Computing, Chicago, IL, USA, June 13-16, 2004, pages 91–100. ACM, 2004. URL: https://doi.org/10.1145/1007352.1007374, doi:10.1145/1007352.1007374.
- [12] Richard Cole, Tsvi Kopelowitz, and Moshe Lewenstein. Suffix trays and suffix trists: Structures for faster text indexing. Algorithmica, 72(2):450–466, 2015. doi:10.1007/s00453-013-9860-6.
- [13] Maxime Crochemore, Christophe Hancart, and Thierry Lecroq. Algorithms on strings. Cambridge University Press, 2007.
- [14] Mark de Berg, Otfried Cheong, Marc J. van Kreveld, and Mark H. Overmars. Computational geometry: algorithms and applications, 3rd Edition. Springer, 2008. URL: https://www.worldcat.org/oclc/227584184.
- [15] Dan F. DeBlasio, Fiyinfoluwa Gbosibo, Carl Kingsford, and Guillaume Marçais. Practical universal k-mer sets for minimizer schemes. In Xinghua Mindy Shi, Michael Buck, Jian Ma, and Pierangelo Veltri, editors, Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, BCB 2019, Niagara Falls, NY, USA, September 7-10, 2019, pages 167–176. ACM, 2019. URL: https://doi.org/10.1145/3307339.3342144, doi:10.1145/3307339.3342144.
- [16] Arthur L. Delcher, Simon Kasif, Robert D. Fleischmann, Jeremy Peterson, Owen White, and Steven L. Salzberg. Alignment of whole genomes. Nucleic Acids Research, 27(11):2369–2376, 01 1999. URL: https://doi.org/10.1093/nar/27.11.2369, arXiv:https://academic.oup.com/nar/article-pdf/27/11/2369/4086173/27-11-2369.pdf, doi:10.1093/nar/27.11.2369.
- [17] Dong Deng, Guoliang Li, and Jianhua Feng. A pivotal prefix based filtering algorithm for string similarity search. In Curtis E. Dyreson, Feifei Li, and M. Tamer Özsu, editors, International Conference on Management of Data, SIGMOD 2014, Snowbird, UT, USA, June 22-27, 2014, pages 673–684. ACM, 2014. URL: https://doi.org/10.1145/2588555.2593675, doi:10.1145/2588555.2593675.
- [18] Dong Deng, Guoliang Li, Jianhua Feng, and Wen-Syan Li. Top-k string similarity search with edit-distance constraints. In Christian S. Jensen, Christopher M. Jermaine, and Xiaofang Zhou, editors, 29th IEEE International Conference on Data Engineering, ICDE 2013, Brisbane, Australia, April 8-12, 2013, pages 925–936. IEEE Computer Society, 2013. URL: https://doi.org/10.1109/ICDE.2013.6544886, doi:10.1109/ICDE.2013.6544886.
- [19] Sebastian Deorowicz, Marek Kokot, Szymon Grabowski, and Agnieszka Debudaj-Grabysz. KMC 2: fast and resource-frugal k-mer counting. Bioinform., 31(10):1569–1576, 2015. URL: https://doi.org/10.1093/bioinformatics/btv022, doi:10.1093/bioinformatics/btv022.
- [20] Patrick Dinklage, Johannes Fischer, Alexander Herlez, Tomasz Kociumaka, and Florian Kurpicz. Practical Performance of Space Efficient Data Structures for Longest Common Extensions. In Fabrizio Grandoni, Grzegorz Herman, and Peter Sanders, editors, 28th Annual European Symposium on Algorithms (ESA 2020), volume 173 of Leibniz International Proceedings in Informatics (LIPIcs), pages 39:1–39:20, Dagstuhl, Germany, 2020. Schloss Dagstuhl–Leibniz-Zentrum für Informatik. URL: https://drops.dagstuhl.de/opus/volltexte/2020/12905, doi:10.4230/LIPIcs.ESA.2020.39.
- [21] Irit Dinur and Shmuel Safra. The importance of being biased. In John H. Reif, editor, Proceedings on 34th Annual ACM Symposium on Theory of Computing, May 19-21, 2002, Montréal, Québec, Canada, pages 33–42. ACM, 2002. URL: https://doi.org/10.1145/509907.509915, doi:10.1145/509907.509915.
- [22] Baris Ekim, Bonnie Berger, and Yaron Orenstein. A randomized parallel algorithm for efficiently finding near-optimal universal hitting sets. In Russell Schwartz, editor, Research in Computational Molecular Biology - 24th Annual International Conference, RECOMB 2020, Padua, Italy, May 10-13, 2020, Proceedings, volume 12074 of Lecture Notes in Computer Science, pages 37–53. Springer, 2020. URL: https://doi.org/10.1007/978-3-030-45257-5_3, doi:10.1007/978-3-030-45257-5\_3.
- [23] Martin Farach. Optimal suffix tree construction with large alphabets. In 38th Annual Symposium on Foundations of Computer Science, FOCS ’97, Miami Beach, Florida, USA, October 19-22, 1997, pages 137–143, 1997. URL: https://doi.org/10.1109/SFCS.1997.646102, doi:10.1109/SFCS.1997.646102.
- [24] Paolo Ferragina and Giovanni Manzini. Indexing compressed text. J. ACM, 52(4):552–581, 2005. doi:10.1145/1082036.1082039.
- [25] Paolo Ferragina and Gonzalo Navarro. Pizza&Chili corpus – compressed indexes and their testbeds. http://pizzachili.dcc.uchile.cl/texts.html.
- [26] Vissarion Fisikopoulos. An implementation of range trees with fractional cascading in C++. CoRR, abs/1103.4521, 2011. URL: http://arxiv.org/abs/1103.4521, arXiv:1103.4521.
- [27] Michael L. Fredman, János Komlós, and Endre Szemerédi. Storing a sparse table with worst case access time. J. ACM, 31(3):538–544, 1984. doi:10.1145/828.1884.
- [28] Travis Gagie, Gonzalo Navarro, and Nicola Prezza. Fully functional suffix trees and optimal text searching in BWT-runs bounded space. J. ACM, 67(1):2:1–2:54, 2020. doi:10.1145/3375890.
- [29] Younan Gao, Meng He, and Yakov Nekrich. Fast preprocessing for optimal orthogonal range reporting and range successor with applications to text indexing. In Fabrizio Grandoni, Grzegorz Herman, and Peter Sanders, editors, 28th Annual European Symposium on Algorithms (ESA 2020), volume 173 of Leibniz International Proceedings in Informatics (LIPIcs), pages 54:1–54:18, Dagstuhl, Germany, 2020. Schloss Dagstuhl–Leibniz-Zentrum für Informatik. URL: https://drops.dagstuhl.de/opus/volltexte/2020/12920, doi:10.4230/LIPIcs.ESA.2020.54.
- [30] Simon Gog, Timo Beller, Alistair Moffat, and Matthias Petri. From theory to practice: Plug and play with succinct data structures. In Joachim Gudmundsson and Jyrki Katajainen, editors, Experimental Algorithms - 13th International Symposium, SEA 2014, Copenhagen, Denmark, June 29 - July 1, 2014. Proceedings, volume 8504 of Lecture Notes in Computer Science, pages 326–337. Springer, 2014. URL: https://doi.org/10.1007/978-3-319-07959-2_28, doi:10.1007/978-3-319-07959-2\_28.
- [31] Szymon Grabowski and Marcin Raniszewski. Sampled suffix array with minimizers. Softw. Pract. Exp., 47(11):1755–1771, 2017. URL: https://doi.org/10.1002/spe.2481, doi:10.1002/spe.2481.
- [32] Roberto Grossi and Jeffrey Scott Vitter. Compressed suffix arrays and suffix trees with applications to text indexing and string matching. SIAM J. Comput., 35(2):378–407, 2005. doi:10.1137/S0097539702402354.
- [33] Wing-Kai Hon, Kunihiko Sadakane, and Wing-Kin Sung. Breaking a time-and-space barrier in constructing full-text indices. SIAM J. Comput., 38(6):2162–2178, 2009. doi:10.1137/070685373.
- [34] Huiqi Hu, Guoliang Li, Zhifeng Bao, Jianhua Feng, Yongwei Wu, Zhiguo Gong, and Yaoqiang Xu. Top-k spatio-textual similarity join. IEEE Trans. Knowl. Data Eng., 28(2):551–565, 2016. URL: https://doi.org/10.1109/TKDE.2015.2485213, doi:10.1109/TKDE.2015.2485213.
- [35] Chirag Jain, Alexander T. Dilthey, Sergey Koren, Srinivas Aluru, and Adam M. Phillippy. A fast approximate algorithm for mapping long reads to large reference databases. J. Comput. Biol., 25(7):766–779, 2018. URL: https://doi.org/10.1089/cmb.2018.0036, doi:10.1089/cmb.2018.0036.
- [36] Chirag Jain, Sergey Koren, Alexander T. Dilthey, Adam M. Phillippy, and Srinivas Aluru. A fast adaptive algorithm for computing whole-genome homology maps. Bioinform., 34(17):i748–i756, 2018. URL: https://doi.org/10.1093/bioinformatics/bty597, doi:10.1093/bioinformatics/bty597.
- [37] Chirag Jain, Arang Rhie, Haowen Zhang, Claudia Chu, Brian Walenz, Sergey Koren, and Adam M. Phillippy. Weighted minimizer sampling improves long read mapping. Bioinform., 36(Supplement-1):i111–i118, 2020. URL: https://doi.org/10.1093/bioinformatics/btaa435, doi:10.1093/bioinformatics/btaa435.
- [38] Tamer Kahveci and Ambuj K. Singh. Efficient index structures for string databases. In Peter M. G. Apers, Paolo Atzeni, Stefano Ceri, Stefano Paraboschi, Kotagiri Ramamohanarao, and Richard T. Snodgrass, editors, VLDB 2001, Proceedings of 27th International Conference on Very Large Data Bases, September 11-14, 2001, Roma, Italy, pages 351–360. Morgan Kaufmann, 2001. URL: http://www.vldb.org/conf/2001/P351.pdf.
- [39] Juha Kärkkäinen, Peter Sanders, and Stefan Burkhardt. Linear work suffix array construction. J. ACM, 53(6):918–936, 2006. doi:10.1145/1217856.1217858.
- [40] Richard M. Karp. Reducibility among combinatorial problems. In Raymond E. Miller and James W. Thatcher, editors, Proceedings of a symposium on the Complexity of Computer Computations, held March 20-22, 1972, at the IBM Thomas J. Watson Research Center, Yorktown Heights, New York, USA, The IBM Research Symposia Series, pages 85–103. Plenum Press, New York, 1972. URL: https://doi.org/10.1007/978-1-4684-2001-2_9, doi:10.1007/978-1-4684-2001-2\_9.
- [41] Richard M. Karp and Michael O. Rabin. Efficient randomized pattern-matching algorithms. IBM J. Res. Dev., 31(2):249–260, 1987. URL: https://doi.org/10.1147/rd.312.0249, doi:10.1147/rd.312.0249.
- [42] Toru Kasai, Gunho Lee, Hiroki Arimura, Setsuo Arikawa, and Kunsoo Park. Linear-time longest-common-prefix computation in suffix arrays and its applications. In Amihood Amir and Gad M. Landau, editors, Combinatorial Pattern Matching, 12th Annual Symposium, CPM 2001 Jerusalem, Israel, July 1-4, 2001 Proceedings, volume 2089 of Lecture Notes in Computer Science, pages 181–192. Springer, 2001. URL: https://doi.org/10.1007/3-540-48194-X_17, doi:10.1007/3-540-48194-X\_17.
- [43] Dominik Kempa and Tomasz Kociumaka. String synchronizing sets: sublinear-time BWT construction and optimal LCE data structure. In Moses Charikar and Edith Cohen, editors, Proceedings of the 51st Annual ACM SIGACT Symposium on Theory of Computing, STOC 2019, Phoenix, AZ, USA, June 23-26, 2019, pages 756–767. ACM, 2019. URL: https://doi.org/10.1145/3313276.3316368, doi:10.1145/3313276.3316368.
- [44] Tomasz Kociumaka. Minimal suffix and rotation of a substring in optimal time. In Roberto Grossi and Moshe Lewenstein, editors, 27th Annual Symposium on Combinatorial Pattern Matching, CPM 2016, June 27-29, 2016, Tel Aviv, Israel, volume 54 of LIPIcs, pages 28:1–28:12. Schloss Dagstuhl - Leibniz-Zentrum für Informatik, 2016. URL: https://doi.org/10.4230/LIPIcs.CPM.2016.28, doi:10.4230/LIPIcs.CPM.2016.28.
- [45] Vladimir I. Levenshtein. Binary codes capable of correcting deletions, insertions and reversals. Soviet Physics Doklady, 10:707, 1966.
- [46] Chen Li, Bin Wang, and Xiaochun Yang. VGRAM: improving performance of approximate queries on string collections using variable-length grams. In Christoph Koch, Johannes Gehrke, Minos N. Garofalakis, Divesh Srivastava, Karl Aberer, Anand Deshpande, Daniela Florescu, Chee Yong Chan, Venkatesh Ganti, Carl-Christian Kanne, Wolfgang Klas, and Erich J. Neuhold, editors, Proceedings of the 33rd International Conference on Very Large Data Bases, University of Vienna, Austria, September 23-27, 2007, pages 303–314. ACM, 2007. URL: http://www.vldb.org/conf/2007/papers/research/p303-li.pdf.
- [47] Heng Li. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics, 32(14):2103–2110, 03 2016. URL: https://doi.org/10.1093/bioinformatics/btw152, arXiv:https://academic.oup.com/bioinformatics/article-pdf/32/14/2103/19567911/btw152.pdf, doi:10.1093/bioinformatics/btw152.
- [48] Heng Li. Minimap2: pairwise alignment for nucleotide sequences. Bioinform., 34(18):3094–3100, 2018. URL: https://doi.org/10.1093/bioinformatics/bty191, doi:10.1093/bioinformatics/bty191.
- [49] Grigorios Loukides and Solon P. Pissis. Bidirectional string anchors: A new string sampling mechanism. In Petra Mutzel, Rasmus Pagh, and Grzegorz Herman, editors, 29th Annual European Symposium on Algorithms, ESA 2021, September 6-8, 2021, Lisbon, Portugal (Virtual Conference), volume 204 of LIPIcs, pages 64:1–64:21. Schloss Dagstuhl - Leibniz-Zentrum für Informatik, 2021. URL: https://doi.org/10.4230/LIPIcs.ESA.2021.64, doi:10.4230/LIPIcs.ESA.2021.64.
- [50] Veli Mäkinen and Gonzalo Navarro. Position-restricted substring searching. In José R. Correa, Alejandro Hevia, and Marcos A. Kiwi, editors, LATIN 2006: Theoretical Informatics, 7th Latin American Symposium, Valdivia, Chile, March 20-24, 2006, Proceedings, volume 3887 of Lecture Notes in Computer Science, pages 703–714. Springer, 2006. URL: https://doi.org/10.1007/11682462_64, doi:10.1007/11682462\_64.
- [51] Udi Manber and Eugene W. Myers. Suffix arrays: A new method for on-line string searches. SIAM J. Comput., 22(5):935–948, 1993. URL: https://doi.org/10.1137/0222058, doi:10.1137/0222058.
- [52] Christopher D. Manning, Prabhakar Raghavan, and Hinrich Schütze. Introduction to Information Retrieval. Cambridge University Press, USA, 2008.
- [53] Guillaume Marçais, Dan F. DeBlasio, and Carl Kingsford. Asymptotically optimal minimizers schemes. Bioinform., 34(13):i13–i22, 2018. URL: https://doi.org/10.1093/bioinformatics/bty258, doi:10.1093/bioinformatics/bty258.
- [54] Guillaume Marçais, David Pellow, Daniel Bork, Yaron Orenstein, Ron Shamir, and Carl Kingsford. Improving the performance of minimizers and winnowing schemes. Bioinform., 33(14):i110–i117, 2017. URL: https://doi.org/10.1093/bioinformatics/btx235, doi:10.1093/bioinformatics/btx235.
- [55] J. Ian Munro, Gonzalo Navarro, and Yakov Nekrich. Space-efficient construction of compressed indexes in deterministic linear time. In Proceedings of the Twenty-Eighth Annual ACM-SIAM Symposium on Discrete Algorithms, SODA 2017, Barcelona, Spain, Hotel Porta Fira, January 16-19, pages 408–424, 2017. doi:10.1137/1.9781611974782.26.
- [56] Yaron Orenstein, David Pellow, Guillaume Marçais, Ron Shamir, and Carl Kingsford. Compact universal k-mer hitting sets. In Martin C. Frith and Christian Nørgaard Storm Pedersen, editors, Algorithms in Bioinformatics - 16th International Workshop, WABI 2016, Aarhus, Denmark, August 22-24, 2016. Proceedings, volume 9838 of Lecture Notes in Computer Science, pages 257–268. Springer, 2016. URL: https://doi.org/10.1007/978-3-319-43681-4_21, doi:10.1007/978-3-319-43681-4\_21.
- [57] Jianbin Qin, Wei Wang, Chuan Xiao, Yifei Lu, Xuemin Lin, and Haixun Wang. Asymmetric signature schemes for efficient exact edit similarity query processing. ACM Trans. Database Syst., 38(3):16:1–16:44, 2013. URL: https://doi.org/10.1145/2508020.2508023, doi:10.1145/2508020.2508023.
- [58] Michael Roberts, Wayne Hayes, Brian R. Hunt, Stephen M. Mount, and James A. Yorke. Reducing storage requirements for biological sequence comparison. Bioinform., 20(18):3363–3369, 2004. URL: https://doi.org/10.1093/bioinformatics/bth408, doi:10.1093/bioinformatics/bth408.
- [59] Craige Schensted. Longest increasing and decreasing subsequences. Canadian Journal of Mathematics, 13:179–191, 1961. doi:10.4153/CJM-1961-015-3.
- [60] Saul Schleimer, Daniel Shawcross Wilkerson, and Alexander Aiken. Winnowing: Local algorithms for document fingerprinting. In Alon Y. Halevy, Zachary G. Ives, and AnHai Doan, editors, Proceedings of the 2003 ACM SIGMOD International Conference on Management of Data, San Diego, California, USA, June 9-12, 2003, pages 76–85. ACM, 2003. URL: https://doi.org/10.1145/872757.872770, doi:10.1145/872757.872770.
- [61] Huei-Jan Shyr and Gabriel Thierrin. Disjunctive languages and codes. In Marek Karpinski, editor, Fundamentals of Computation Theory, Proceedings of the 1977 International FCT-Conference, Poznan-Kórnik, Poland, September 19-23, 1977, volume 56 of Lecture Notes in Computer Science, pages 171–176. Springer, 1977. URL: https://doi.org/10.1007/3-540-08442-8_83, doi:10.1007/3-540-08442-8\_83.
- [62] Yihan Sun and Guy E. Blelloch. Parallel range, segment and rectangle queries with augmented maps. In Stephen G. Kobourov and Henning Meyerhenke, editors, Proceedings of the Twenty-First Workshop on Algorithm Engineering and Experiments, ALENEX 2019, San Diego, CA, USA, January 7-8, 2019, pages 159–173. SIAM, 2019. URL: https://doi.org/10.1137/1.9781611975499.13, doi:10.1137/1.9781611975499.13.
- [63] The CGAL Project. CGAL User and Reference Manual. CGAL Editorial Board, 5.2.1 edition, 2021. URL: https://doc.cgal.org/5.2.1/Manual/packages.html.
- [64] Jiannan Wang, Guoliang Li, and Jianhua Feng. Can we beat the prefix filtering?: an adaptive framework for similarity join and search. In K. Selçuk Candan, Yi Chen, Richard T. Snodgrass, Luis Gravano, and Ariel Fuxman, editors, Proceedings of the ACM SIGMOD International Conference on Management of Data, SIGMOD 2012, Scottsdale, AZ, USA, May 20-24, 2012, pages 85–96. ACM, 2012. URL: https://doi.org/10.1145/2213836.2213847, doi:10.1145/2213836.2213847.
- [65] Xiaoli Wang, Xiaofeng Ding, Anthony K. H. Tung, and Zhenjie Zhang. Efficient and effective KNN sequence search with approximate n-grams. Proc. VLDB Endow., 7(1):1–12, 2013. URL: http://www.vldb.org/pvldb/vol7/p1-wang.pdf, doi:10.14778/2732219.2732220.
- [66] Peter Weiner. Linear pattern matching algorithms. In 14th Annual Symposium on Switching and Automata Theory, Iowa City, Iowa, USA, October 15-17, 1973, pages 1–11, 1973. doi:10.1109/SWAT.1973.13.
- [67] Derrick E. Wood and Steven L. Salzberg. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biology, 15(3), March 2014. Copyright: Copyright 2014 Elsevier B.V., All rights reserved. doi:10.1186/gb-2014-15-3-r46.
- [68] Zhenglu Yang, Jianjun Yu, and Masaru Kitsuregawa. Fast algorithms for top-k approximate string matching. In Maria Fox and David Poole, editors, Proceedings of the Twenty-Fourth AAAI Conference on Artificial Intelligence, AAAI 2010, Atlanta, Georgia, USA, July 11-15, 2010. AAAI Press, 2010. URL: http://www.aaai.org/ocs/index.php/AAAI/AAAI10/paper/view/1939.
- [69] Minghe Yu, Jin Wang, Guoliang Li, Yong Zhang, Dong Deng, and Jianhua Feng. A unified framework for string similarity search with edit-distance constraint. VLDB J., 26(2):249–274, 2017. URL: https://doi.org/10.1007/s00778-016-0449-y, doi:10.1007/s00778-016-0449-y.
- [70] Haoyu Zhang and Qin Zhang. Minsearch: An efficient algorithm for similarity search under edit distance. In Rajesh Gupta, Yan Liu, Jiliang Tang, and B. Aditya Prakash, editors, KDD ’20: The 26th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, Virtual Event, CA, USA, August 23-27, 2020, pages 566–576. ACM, 2020. URL: https://doi.org/10.1145/3394486.3403099, doi:10.1145/3394486.3403099.
- [71] Zhenjie Zhang, Marios Hadjieleftheriou, Beng Chin Ooi, and Divesh Srivastava. Bed-tree: an all-purpose index structure for string similarity search based on edit distance. In Ahmed K. Elmagarmid and Divyakant Agrawal, editors, Proceedings of the ACM SIGMOD International Conference on Management of Data, SIGMOD 2010, Indianapolis, Indiana, USA, June 6-10, 2010, pages 915–926. ACM, 2010. URL: https://doi.org/10.1145/1807167.1807266, doi:10.1145/1807167.1807266.
- [72] Hongyu Zheng, Carl Kingsford, and Guillaume Marçais. Improved design and analysis of practical minimizers. Bioinform., 36(Supplement-1):i119–i127, 2020. URL: https://doi.org/10.1093/bioinformatics/btaa472, doi:10.1093/bioinformatics/btaa472.
- [73] Hongyu Zheng, Carl Kingsford, and Guillaume Marçais. Lower density selection schemes via small universal hitting sets with short remaining path length. In Russell Schwartz, editor, Research in Computational Molecular Biology - 24th Annual International Conference, RECOMB 2020, Padua, Italy, May 10-13, 2020, Proceedings, volume 12074 of Lecture Notes in Computer Science, pages 202–217. Springer, 2020. URL: https://doi.org/10.1007/978-3-030-45257-5_13, doi:10.1007/978-3-030-45257-5\_13.
- [74] Hongyu Zheng, Carl Kingsford, and Guillaume Marçais. Sequence-specific minimizers via polar sets. bioRxiv, 2021. URL: https://www.biorxiv.org/content/early/2021/02/10/2021.02.01.429246, arXiv:https://www.biorxiv.org/content/early/2021/02/10/2021.02.01.429246.full.pdf, doi:10.1101/2021.02.01.429246.