Source-Free Collaborative Domain Adaptation via Multi-Perspective Feature Enrichment for Functional MRI Analysis
Abstract
Resting-state functional MRI (rs-fMRI) is increasingly employed in multi-site research to aid neurological disorder analysis. Existing studies usually suffer from significant cross-site/domain data heterogeneity caused by site effects such as differences in scanners/protocols. Many methods have been proposed to reduce fMRI heterogeneity between source and target domains, heavily relying on the availability of source data. But acquiring source data is challenging due to privacy concerns and/or data storage burdens in multi-site studies. To this end, we design a source-free collaborative domain adaptation (SCDA) framework for fMRI analysis, where only a pretrained source model and unlabeled target data are accessible. Specifically, a multi-perspective feature enrichment method (MFE) is developed for target fMRI analysis, consisting of multiple collaborative branches to dynamically capture fMRI features of unlabeled target data from multiple views. Each branch has a data-feeding module, a spatiotemporal feature encoder, and a class predictor. A mutual-consistency constraint is designed to encourage pair-wise consistency of latent features of the same input generated from these branches for robust representation learning. To facilitate efficient cross-domain knowledge transfer without source data, we initialize MFE using parameters of a pretrained source model. We also introduce an unsupervised pretraining strategy using 3,806 unlabeled fMRIs from three large-scale auxiliary databases, aiming to obtain a general feature encoder. Experimental results on three public datasets and one private dataset demonstrate the efficacy of our method in cross-scanner and cross-study prediction tasks. The model pretrained on large-scale rs-fMRI data has been released to the public.
Index Terms:
Source-free domain adaptation, feature enrichment, collaborative learning, unsupervised pretraining.I Introduction
Neurological disorders include a broad range of brain diseases, such as autism spectrum disorder and cognitive impairment [1, 2], caused by a combination of genetic, environmental, and psychological factors. The treatments for these brain diseases emerge challenges to health care systems globally, bringing society much economic and manpower burden. Therefore, early and accurate identification of these disorders is much critical to helping clinicians develop timely interventions. Resting-state functional MRI (rs-fMRI) provides a non-invasive technique to measure spontaneous brain activity through blood-oxygen-level-dependent (BOLD) signals [3]. It has been increasingly used to study functional brain systems and has shown great clinical value in neurological disorder analysis [4].

Many studies [5, 6] employ multi-site fMRI to increase sample size for brain disorder identification, but many of them neglect significant cross-site/domain data heterogeneity caused by site effects such as differences in scanner vendors and/or imaging protocols [7], leading to suboptimal learning performance. Several recent studies [8, 9] explore unsupervised domain adaptation (see Fig. 1 (a)) to reduce fMRI heterogeneity between source and target domains. They generally heavily rely on the availability of source data for target model construction. For instance, Gao et al. [9] propose to use labeled source fMRI to generate a distinct descriptor, and then minimize the discrepancy between source and target distributions to achieve cross-subject decoding. Huang et al. [8] also rely on labeled source fMRI to learn a discriminative fMRI representation, and then they perform cross-domain adaptation using an adversarial-based neural network. However, direct accessing source data is usually challenging in multi-site studies due to privacy concerns and/or data storage burdens. Medical data may contain subjects’ personal information, and directly accessing these data may cause privacy leakage [10]. Additionally, multi-site studies often have a large amount of data, which may cause a data storage burden [11].
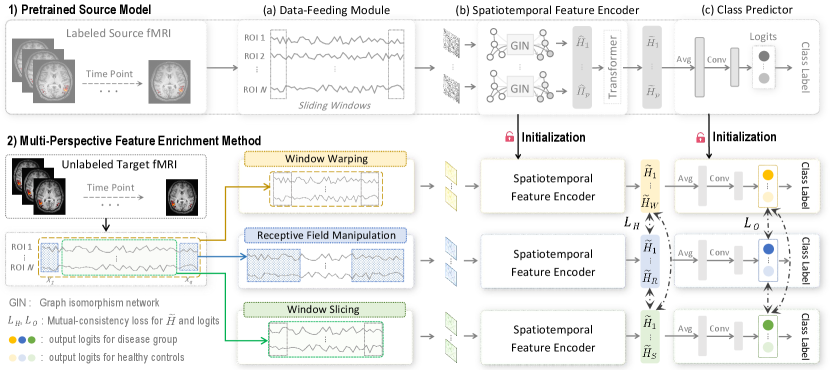
To this end, we propose a novel source-free collaborative domain adaptation (SCDA) framework for fMRI analysis and brain disorder diagnosis, without accessing any source data for target model training. This study aims to learn a target model for improving adaptation performance only based on a pretrained source model and unlabeled target fMRI data (see Fig. 1 (b)). Specifically, we design a multi-perspective feature enrichment method (MFE), which consists of multiple collaborative branches to dynamically exploit unlabeled target data from multiple perspectives, e.g., window warping, receptive field manipulation, and window slicing. Each branch of the MFE comprises a data-feeding module, a spatiotemporal feature encoder, and a class predictor. A mutual-consistency constraint is also designed to enforce pair-wise consistency of latent features generated by these branches for robust representation learning of unlabeled target data. To facilitate source-to-target knowledge transfer without accessing source data, we initialize the MFE using parameters of a pretrained source model. To obtain a general fMRI encoder, we further propose an effective unsupervised pretraining strategy based on 3,806 unlabeled fMRI data from three large-scale auxiliary fMRI databases, i.e., ABIDE [12], REST-meta-MDD [13], and ADHD-200 [14]. We evaluate our proposed SCDA on 1,591 rs-fMRI scans from three public datasets and one private dataset, with experimental results suggesting its efficacy in both cross-scanner and cross-study prediction tasks. And the identified discriminative brain regions can be used as potential biomarkers to assist fMRI-based brain disorder analysis. The contributions of this work are summarized below.
-
•
A novel source-free collaborative domain adaptation (SCDA) framework is designed for fMRI-based neurological disorder diagnosis without accessing source data. To our knowledge, this is among the first attempts to investigate source-free domain adaptation in fMRI analysis.
-
•
A multi-perspective feature enrichment method is designed to exploit dynamic characteristics of target data from multiple views. It can be straightforwardly applied to other applications for multi-view feature learning.
-
•
An unsupervised pretraining strategy is proposed to leverage large-scale auxiliary unlabeled fMRI databases to generate a general feature encoder, which helps learn more transferable fMRI features and enhances effective cross-domain adaptation. The model pretrained on large-scale rs-fMRI data has been shared with the public111https://github.com/yqfang9199/SCDA.
-
•
Comprehensive experiments in cross-scanner and cross-study prediction tasks using three public datasets and one private dataset demonstrate the effectiveness of the SCDA. And the disorder-associated brain regions identified by the SCDA could be used as potential biomarkers to facilitate fMRI-based neurological disorder analysis.
The remainder of this paper is organized as follows. Section II reviews the most relevant studies on fMRI adaptation and fMRI representation learning. Section III introduces details of the proposed method. Section IV presents materials and results. Section V discusses the influence of key components of our method. The paper is concluded in Section VI.
II Related Work
II-A Functional MRI Adaptation
Functional MRI is increasingly employed to study brain systems and has shown great clinical value in neurological disorder diagnosis [15, 16]. To increase sample size and enhance statistical power, many studies [5, 6] propose to leverage multi-site/domain fMRI data for brain disorder analysis. These studies usually assume that fMRI data are sampled from the same distribution while neglecting significant inter-domain heterogeneity caused for instance by the use of different scanners/protocols, which could lead to biased or inaccurate predictions. To address this domain distribution shift problem, many fMRI adaptation approaches [17, 18, 19, 9] have been proposed to perform data/feature harmonization across domains, yielding robust prediction models to improve inference in target domain. Even with improved prediction performance, these studies heavily rely on the availability of source data for source-to-target adaptation. Zhang et al. [18] present a multi-way adaptation architecture, which performs joint distribution alignment in each pair of source and target fMRI based on optimal transport. Gao et al. [9] propose to use source fMRI to derive a distinct descriptor, and then they perform cross-subject adaptation by reducing discrepancy between source and target distributions. But acquiring source data is usually challenging due to privacy concerns and/or data storage burdens, while existing studies seldom investigate fMRI adaptation without accessing source data. To bridge this gap, we propose a source-free collaborative domain adaptation method, where only a pretrained source model and unlabeled target data are available for target model construction. We aim to transfer knowledge from a pretrained source model to unlabeled target data to improve target inference.
II-B Functional MRI Representation Learning
Representation learning aims to extract informative features from fMRI data, which can facilitate subsequent fMRI analysis and interpretation of brain activity. Existing studies for fMRI representation learning can be mainly categorized into traditional machine learning methods and deep learning based methods. The traditional methods usually extract handcrafted fMRI features (e.g., clustering coefficient, modularity, and density) for downstream prediction [20], and the commonly used prediction models include support vector machine (SVMs) [20], eXtreme Gradient Boosting (XGBoost) [21], and random forest (RF) [22]. In these methods, the extracted human-engineered features require much expert knowledge, which may not be optimal for subsequent prediction.
To tackle the problem, many deep learning methods have been developed to automatically learn task-oriented fMRI features [23, 24, 25, 26, 27], which usually show superior performance compared with traditional machine learning models. For example, Li et al. [24] propose an interpretable graph neural network, which leverages both topological and functional characteristics of fMRI data for neurological disorder diagnosis. Kim et al. [25] introduce an attention-based neural network to incorporate both spatial and temporal information of fMRI data for brain connectome analysis. Among existing fMRI studies, we find they usually leverage original fMRI timeseries directly, without considering multi-perspective representations of fMRI. For example, some works [28, 29] use sliding window strategy to characterize temporal dynamics of fMRI data, but they usually employ fixed window size, limiting the ability to capture brain temporal variability from various receptive fields. In this work, we propose to dynamically exploit fMRI data from multiple perspectives, e.g., window warping, receptive field manipulation, and window slicing, which helps capture multi-view informative fMRI features. Furthermore, our method introduces an unsupervised pretraining strategy, by training our model on 3,806 subjects from three large-scale auxiliary fMRI databases. Benefiting from these massive and diverse data, it is expected to capture more general fMRI features for improving performance in downstream tasks.
III Methodology
To protect source data privacy and reduce data storage burden in multi-site studies, we develop a source-free collaborative domain adaptation (SCDA) framework for fMRI analysis, where only a pretrained source model and unlabeled target data are accessible for target model construction. As shown in Fig. 2, the SCDA consists of 1) a pretrained source model for discriminative fMRI feature extraction, and 2) a multi-perspective feature enrichment method (MFE) for target inference using unlabeled target data from various views. The MFE is initialized using parameters of the pretrained source model for cross-domain knowledge transfer, and optimized via a mutual-consistency constraint. We also propose an unsupervised pretraining strategy to obtain a general fMRI feature encoder, which aims to facilitate cross-domain adaptation.
III-A Construction of Pretrained Source Model
A pretrained source model is used to facilitate source-to-target knowledge transfer. As shown in the top of Fig. 2, the source model takes full-length fMRI timeseries as input and consists of a data-feeding module, a spatiotemporal feature encoder, and a class predictor, elaborated as follows.
III-A1 Data-Feeding Module
In this module, each subject is represented by its fMRI timeseries , where and denote the number of time points and regions-of-interest (ROIs), respectively. To characterize brain functional variability across time, we partition into multiple overlapping sliding windows, and construct windowed timeseries based on a sliding window with the length of and the stride of , where is floor function. Here, we represent each windowed timeseries as a functional brain network/graph, where graph nodes indicate brain ROIs and edges represent functional connections among these ROIs. Denote , , and as the vertex set, the edge set, and the adjacency matrix, respectively. At the -th sliding window, we represent as a graph , where each node represents the -th ROI with its feature vector and each edge represents the Pearson’s correlation coefficient (PCC) between the -th and the -th ROIs. The adjacency matrix is derived by keeping the top 30% strongest edges of each graph, where denotes there exists an edge between two ROIs while denotes there is no edge.
III-A2 Spatiotemporal Feature Encoder
With a set of graphs for each subject as input, a spatiotemporal feature encoder is designed to learn fMRI features from spatial and temporal aspects. Specifically, we introduce 1) a graph neural network for spatial feature aggregation across ROIs and 2) a Transformer for temporal attention learning across sliding windows, which are detailed as follows.
1) Spatial Feature Aggregation. Graph neural networks (GNNs) typically update node representations by aggregating features of their neighbors iteratively, thereby capturing spatial/structural information across the entire graph [24, 30]. We employ graph isomorphism network (GIN) [31] for spatial representation learning, a GNN variant with high discriminative power in graph classification. Mathematically, the node-level embedding at the -th sliding window, formulated as:
| (1) |
where denotes the feature vector of node at layer , is one-hot encoding of node , is a multiple-layer perceptron, is a learnable parameter initialized with 0, and denotes neighbors of node . Following [25], with , we can formulate Eq. (1) as:
| (2) |
where denotes all node features for the -th layer at sliding window , is an identity matrix, is an adjacent matrix, means weights, and is a nonlinear function. By concatenating node features of all layers, we obtain a new node-level embedding at sliding window . We further embed a squeeze-excitation block [32] into to automatically locate informative ROIs, where a spatial attention vector is represented as . Here, calculates the mean of , and are learnable weight matrices, is an activation function with batch normalization [33] and Gaussian error linear unit [34]. Each element in denotes the importance of a specific ROI. The spatial-attended features () can be obtained by multiplying and , followed by an average operation across ROIs.
2) Temporal Attention Learning. With spatial-attended features of sliding windows for each subject, we also capture temporal attention across different windows. Denote , , and as linear operations. Specifically, we model such temporal attention using a single-head Transformer [35] based on a self-attention matrix , where , , and is a scaling factor for computation stability. Then, we can derive spatiotemporally-attended feature by multiplying and , followed by an for feature abstraction.
III-A3 Class Predictor
To obtain a whole-graph embedding for each subject, we average the spatiotemporal fMRI features across all sliding windows. The derived embedding is fed into a linear fully-connected layer to output a prediction logit . With the generated logits and ground-truth labels for all source samples, a cross-entropy loss function is used to train the source model in a supervised manner. In this way, we can obtain a pretrained source model for downstream target tasks, and the source data and labels are not available for target model construction.
III-B Multi-Perspective Feature Enrichment
To perform source-free domain adaptation with a pretrained source model and unlabeled target data, we propose a multi-perspective feature enrichment method (MFE). As shown in the bottom of Fig. 2, it consists of multiple collaborative branches to dynamically capture target fMRI features from three views (i.e., window warping, receptive field manipulation, and window slicing). Similar to the source model, each branch of MFE is equipped with a data-feeding module, a spatiotemporal feature encoder, and a class predictor. We initialize the MFE using parameters of the pretrained source model to facilitate source-to-target knowledge transfer without accessing source data.
III-B1 Feature Enrichment Strategy
The data-feeding module of each branch corresponds to one feature enrichment perspective, and the feature encoder and class predictor share similar network architecture as the source model. The three feature enrichment strategies used for fMRI representation learning are introduced as follows.
(1) Window Warping. The window warping is an augmentation strategy to stretch or contract a timeseries [36]. In our work, given an fMRI timeseries, we perform window warping on it with a resampling ratio within the domain of , where corresponds to an upsampling operation that increases the temporal resolution of fMRI timeseries, corresponds to a downsampling operation that decreases the temporal resolution of fMRI timeseries, and denotes that the original timeseries is retained. All resampling operations for fMRI data are performed based on fast Fourier transform (FFT) algorithm [37]. During the target model training process, we randomly select different and dynamically generate resampled fMRI timeseries. We also leverage an overlapping sliding window strategy (with length and stride ) to partition the resampled timeseries, and the derived windowed timeseries serve as the input of the spatiotemporal feature encoder.
The rationales that leverage window warping are as follows. (a) By upsampling the fMRI timeseries, we can obtain more data points and capture more rapid changes in brain activity, which helps the feature encoder capture fine-grained fMRI patterns. (b) By downsampling the fMRI timeseries, we can reduce the number of data points and capture coarse-grained fMRI patterns and more general temporal trends. This practice may filter out random noise and make the timeseries smoother, which helps improve the signal-to-noise ratio, thus making the feature encoder capture more informative fMRI patterns. (c) In fMRI domain adaptation, source and target data are usually collected from different scanners/protocols, which may have different temporal resolutions. This window warping strategy can help the feature encoder capture fMRI timeseries with varying temporal resolutions, catering to different image acquisition settings in multi-site studies.
(2) Receptive Field Manipulation. Many fMRI studies [28, 29] leverage the sliding window scheme to characterize temporal dynamics of fMRI. To further model brain variability across time, we design a unique receptive field manipulation strategy to dynamically adjust the size of sliding windows. Specifically, we randomly select the window size within the range of during training, and the derived multi-scale windowed timeseries are fed into the spatiotemporal feature encoder for fMRI feature learning.
The rationales that we dynamically manipulate window sizes are as follows. Using different sizes of sliding windows enables the examination of brain activity from different scales. Specifically, using a larger window size (e.g., 100) that covers a longer timeseries span helps the feature encoder capture more contextual fMRI patterns. Using a smaller window size (e.g., 40) can help the feature encoder detect more transient brain changes and finer-grained temporal variations. The combination of both of them may provide a more comprehensive representation of brain variability inherent in fMRI signals.
(3) Window Slicing. Existing studies mainly use full-length fMRI timeseries for feature extraction while ignoring augmenting timeseries by generating multiple fMRI segments from a full-length one. We dynamically slice fMRI timeseries from the full-length one based on a ratio , and these sliced timeseries serve as the input of the spatiotemporal feature encoder. Here, , where and mean the length (i.e., time points) of sliced timeseries and full-length timeseries of target fMRI. At each training step, the sliced fMRI timeseries change dynamically, i.e., each with a different starting point and a different length. Most fMRI studies usually have only a limited sample size (e.g., tens), which negatively affects the capability of the learned model. So in this work, we propose to generate more fMRI segments based on window slicing to augment input samples, encouraging the feature encoder to learn more robust fMRI patterns to improve learning performance.
In MFE, each branch corresponds to a specific feature enrichment strategy. We initialize each branch using parameters of the pretrained source model to facilitate cross-domain knowledge transfer (see Fig. 2). In this way, we can transfer discriminative fMRI representations learned from source domain to target domain, which helps yield a robust target model for improved inference based on unlabeled target data.
III-B2 Mutual-Consistency Constraint
As mentioned above, to fully exploit unlabeled target fMRI data, we develop a feature enrichment strategy to generate multi-perspective features for each input subject. Intuitively, these features tend to be similar, because they are generated from the same subject. Accordingly, we design a mutual-consistency constraint to train the MFE for target inference. The main idea is to encourage fMRI features generated from various perspectives to be consistent. Mathematically, we propose to enforce pair-wise consistency of spatiotemporal features and output logits generated by three branches by minimizing the following mutual-consistency constraint:
| (3) |
| (4) |
| (5) |
where and denotes spatiotemporal features and output logits generated from -th feature enrichment strategy for -th subject, respectively. Here, denotes the number of feature enrichment strategies ( in this work), denotes the number of target subjects. With Eq. (3), three branches of MFE (initialized by the pretrained source model) can be collaboratively fine-tuned on unlabeled target data, thus catering to the data distribution of the target domain.
III-B3 Inference
During inference, given a new test subject with fMRI timeseries, we first employ the sliding window scheme for timeseries partition, and the windowed timeseries are then fed into each branch of MFE to generate prediction logits. The logits are then converted to predicted probabilities based on a function, and we average the probabilities of three branches to derive final predictions.
III-C Unsupervised Pretraining on Large-Scale Auxiliary fMRI
While there may be limited fMRI data in the source domain, there exist many large-scale auxiliary fMRI databases (even without task-specific label information) that can be employed to facilitate fMRI feature extraction. To this end, we propose an unsupervised pretraining strategy by using three large-scale fMRI databases, including ABIDE [12], REST-meta-MDD [13], and ADHD-200 [14]. From the three databases, we select subjects with fMRI length (i.e., time points) exceeding 170, resulting in 3,806 auxiliary rs-fMRI scans. The demographic information of these subjects is shown in Table I, and their ID information is listed in Supplementary Materials222The diagnostic labels of 26 subjects in Brown site in ADHD-200 are not provided. The demographic information for 48 subjects of Site-4 in REST-meta-MDD is not provided. The gender of one subject at NYU2 (ID: 10044) in ADHD-200 is not given. 180 subjects in ADHD-200 have two fMRI scans..
| Database | ABIDE | REST-meta-MDD | ADHD-200 | |||||
|---|---|---|---|---|---|---|---|---|
| ASD | HC | MDD | HC | ADHD | HC | |||
| Subject # | 351 | 370 | 1,163 | 1,004 | 285 | 379 | ||
| Gender (M/F) | 308/43 | 295/75 | 415/748 | 425/579 | 232/52 | 196/183 | ||
| Age (meanstd) | 16.9±8.0 | 16.6±6.8 | 36.9±14.9 | 37.0±16.0 | 12.2±3.1 | 13.2±3.5 | ||
In our unsupervised pretraining strategy, we leverage the same network architecture as MFE introduced in Section III-B, and use these auxiliary fMRI data as input for model pretraining in an unsupervised manner. The pretrained MFE will be used to initialize the source model. Since these auxiliary fMRI data are acquired from multi-site studies that use different scanners and/or scanning protocols, and even from different diseases and populations, this pretraining strategy is expected to help produce a general fMRI feature encoder. Accordingly, features learned from this general feature encoder will be less affected by domain bias and more transferable to downstream tasks, helping address cross-domain shift problem. Our pretrained model has been released to the public333https://github.com/yqfang9199/SCDA.
The proposed SCDA is implemented via PyTorch with Adam optimizer (mini-batch size: 64). The initial learning rate is set to 0.0003 and dropped by 0.5 every 50 epochs, with a total of 150 training epochs. The experiments are performed on a GPU (NVIDIA TITAN Xp) with 12GB of memory.
| Group | Cross-scanner Prediction with ABIDE | Cross-scanner Prediction with ADHD-200 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NYU1 (source domain) | UM (target domain) | NYU2 (source domain) | Peking (target domain) | ||||||||
| ASD | HC | ASD | HC | ADHD | HC | ADHD | HC | ||||
| Subject No. | 75 | 100 | 66 | 74 | 152 | 111 | 102 | 143 | |||
| Gender (M/F) | 65/10 | 74/26 | 57/9 | 56/18 | 115/36 | 55/56 | 90/12 | 84/59 | |||
| Age (mean±std) | 14.74±7.08 | 15.65±6.16 | 13.17±2.40 | 14.81±3.61 | 10.97±2.67 | 12.11±3.10 | 12.09±2.04 | 11.43±1.86 | |||
| Time points () | 176 | 176 | 296 | 296 | 171 | 171 | 231 | 231 | |||
| Group | Cross-study Prediction with REST-meta-MDD and ABIDE | Cross-study Prediction with ADHD-200 and T2DM | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Site-21 (source domain) | NYU1 (target domain) | NYU2 (source domain) | T2DM (target domain) | ||||||||
| MDD | HC | ASD | HC | ADHD | HC | T2DM-CI | HC | ||||
| Subject No. | 282 | 251 | 75 | 100 | 152 | 111 | 30 | 31 | |||
| Gender (M/F) | 99/183 | 87/164 | 65/10 | 74/26 | 115/36 | 55/56 | 20/10 | 21/10 | |||
| Age (mean±std) | 38.74±13.74 | 39.64±15.87 | 14.74±7.08 | 15.65±6.16 | 10.97±2.67 | 12.11±3.10 | 52.15±8.29 | 50.48±7.07 | |||
| Time points () | 232 | 232 | 176 | 176 | 171 | 171 | 950 | 950 | |||
IV Experiments
IV-A Materials
IV-A1 Data Acquisition and Preprocessing
Three public datasets and one private dataset are employed in this work. 1) ABIDE [12]: This dataset contains 1,112 rs-fMRI scans acquired from 16 international imaging sites, including 539 subjects diagnosed with autism spectrum disorder (ASD) and 573 healthy controls (HCs). 2) REST-meta-MDD [13]: This database contains 2,428 rs-fMRI data acquired from 25 research groups, including 1,300 subjects suffering from major depressive disorder (MDD) and 1,128 HCs. 3) ADHD-200 [14]: This dataset is a collaboration of 8 imaging sites with 1,395 rs-fMRI scans acquired from 388 subjects diagnosed with attention-deficit/hyperactivity disorder (ADHD) and 585 HCs. 4) T2DM: This rs-fMRI dataset is collected from The First Affiliated Hospital of Guangzhou University of Chinese Medicine, which aims to investigate Type 2 diabetes mellitus-associated cognitive impairment. Specifically, a Siemens (Munich, Germany) 3T Prisma scanner with a 64-channel head coil is used to acquire fMRI scans. The gradient echo-planar imaging sequence acquisition parameters are as follows: repetition time (TR) is ; echo time (TE) is ; field-of-view is ; slice thickness is ; voxel size is . T2DM consists of 30 subjects with cognitive impairment (T2DM-CI) and 31 HCs.
A total of 3,806 auxiliary rs-fMRI scans from three datasets (i.e., ABIDE, REST-meta-MDD, and ADHD-200) are used for unsupervised pretraining, where no label information of these data is used. These auxiliary fMRIs are used to construct an initial MFE to obtain a general encoder, and the learned network parameters are used to initialize the source model.
The rs-fMRIs are preprocessed using a standard pipeline, with several key steps listed as follows: 1) magnetization equilibrium, 2) slice timing correction, 3) head motion correction, 4) bandpass filtering, 5) nuisance signal removal, 6) registration between T1-weighted MRI and mean functional images, 7) spatial normalization to Montreal Neurological Institute (MNI) template, and 8) brain partition into =116 ROIs based on automated anatomical atlas (AAL). We use regional mean timeseries to represent each subject.
IV-A2 Domain Setting
We perform cross-scanner and cross-study prediction to validate the efficacy of SCDA in cross-domain fMRI adaptation, where domain denotes scanner and study, respectively. The demographic characteristics of the studied subjects in cross-scanner and cross-study prediction are given in Table II and Table III, respectively.
1) Cross-scanner prediction. In multi-site studies, different hospitals/sites may use different scanners and imaging protocols to acquire fMRI data, leading to data distribution differences across sites. These differences may negatively affect the performance and generalizability of models learned from one site when applied to another site. Cross-scanner prediction is essential to reduce distribution shifts between sites. Here, we conduct two groups of experiments to investigate cross-scanner prediction: 1) Two largest sites (i.e., NYU1 and UM) of ABIDE are used to identify ASD patients from HCs, where NYU1 with 175 subjects serves as the source domain and UM with 140 subjects is used as the target domain. Data of NYU1 and UM are collected by using a Siemens Magnetom Allegra MRI scanner and a GE Signa MRI scanner, respectively. 2) Two largest sites (i.e., NYU2 and Peking) of ADHD-200 are used to identify ADHD patients from HCs, where NYU2 with 263 subjects is employed as the source domain and Peking with 245 subjects serves as the target domain. Data of NYU2 and Peking are collected by using a Siemens Magnetom Allegra MRI scanner and a Siemens Magnetom Trio Tim MRI scanner, respectively.
2) Cross-study prediction. Many studies have limited sample sizes due to various reasons such as disease rarity and patient dropout, while some other studies may have relatively large data. Cross-study prediction allows for leveraging labeled data from one study to assist in learning models for another study with limited sample sizes. Here, we conduct two groups of experiments to investigate cross-study prediction: 1) The largest site (i.e., Site-21, with 533 subjects) of REST-meta-MDD is used as the source domain and the largest site (i.e., NYU1, with 175 subjects) of ABIDE serves as the target domain. 2) The largest site (i.e., NYU2, with 263 subjects) of ADHD-200 is used as the source domain and the T2DM (with 61 subjects) is employed as the target domain.
IV-B Competing Methods
We compare our proposed SCDA with five traditional methods, including 1) NF-SVM, 2) GF-SVM, 3) PCC-XGB, 4) PCC-RF, 5) MaLRR [17], and six deep learning methods, including 6) DANN [38], 7) RAINCOAT [39], 8) UFA-Net [19], 9) SHOT [40], 10) CDCL [41], and 11) SCDA-Naive, with details introduced as follows.
1) NF-SVM: In this method, we first construct a functional connectivity (FC) matrix for each subject based on PCC which measures linear correlation between paired ROIs. Then, we extract six node-based features (i.e., clustering coefficient, node strength, local efficiency, eigenvector centrality, modularity, and node betweenness centrality) from the FC matrix and perform prediction using a support vector machine (SVM).
2) GF-SVM: Based on the same FC matrix in NF-SVM, this method extracts five graph-based features (i.e., density, global efficiency, assortativity coefficient, characteristic path length, and transitivity) and performs prediction using an SVM.
3) PCC-XGB: In this method, we first build an FC matrix based on PCC, and then we flatten the upper triangle elements of the FC matrix and convert them into a one-dimensional vectorized representation. An XGBoost model [42] is then used to produce prediction results.
4) PCC-RF: This method uses the same features as PCC-XGB but with a random forest classifier for final prediction.
5) MaLRR: It tackles multi-site domain adaptation based on rs-fMRI signals via low-rank representation (LRR) decomposition. Specifically, it first captures spatiotemporal fMRI features of source and target domains and transforms them into a common feature space via LRR, and then each source sample is linearly represented using all target samples. The transformed source and target features are then fed into an SVM model for final classification.
6) DANN: This is a widely used unsupervised domain adaptation method. For a fair comparison, we replace the original feature encoder with our spatiotemporal feature encoder and randomly capture fMRI features from various perspectives (same as our SCDA). A domain discriminator is then used to differentiate which features come from which domain, thereby encouraging domain invariance in feature space.
7) RAINCOAT: This recently proposed method is designed to tackle domain adaptation for timeseries data. Specifically, RAINCOAT takes advantage of both time and frequency characteristics during feature encoding in order to exploit time-frequency features of timeseries data. With these encoded features, a Sinkhorn divergence is then leveraged to perform feature alignment between source and target domains.
8) UFA-Net: This method is specifically designed to address unsupervised domain adaptation for rs-fMRI data. Specifically, it employs an attention-based graph convolutional network to capture fMRI representations for both source and target domains, and then a discrepancy-constrained module is introduced to align cross-domain feature representations.
9) SHOT: This method performs source-free domain adaptation only using a pretrained source model and unlabeled target data (same as our SCDA). Specifically, SHOT leverages knowledge stored in the source model to exploit target-specific representations via both information maximization and clustering-based pseudo-labeling. Similar to SCDA, this method leverages unsupervised pretraining based on the three auxiliary fMRI databases to initialize the source model.
10) CDCL: This method also proposes a source-free domain adaptation strategy. Specifically, CDCL first generates source prototypical features from a pretrained source model to represent cluster centers of different categories. Then, a contrastive self-supervised learning framework is designed for cross-domain feature alignment, which aims to pull features of similar categories closer and push features of different categories away in feature space. Similarly, we also utilize unsupervised pretraining based on three auxiliary fMRI databases for source model initialization.
11) SCDA-Naive: It represents the SCDA without unsupervised pretraining, i.e., the source model is trained from scratch rather than initialized from large-scale fMRI databases.
Note that the four traditional methods (i.e., NF-SVM, GF-SVM, PCC-XGB, PCC-RF) only access source data for model training, while the learned models are directly applied to the target domain during inference. The four unsupervised domain adaptation methods (i.e., MaLRR, DANN, RAINCOAT, UFA-Net) need to access labeled source data and unlabeled target data for target model construction. The four source-free unsupervised domain adaptation methods (i.e., SHOT, CDCL, SCDA-Naive, and SCDA) only use a pretrained source model and unlabeled target data for target model learning. All deep learning methods are repeated five times independently to reduce the bias caused by parameter initialization.
| Method | Task 1: ASD vs. HC classification on UM | Task 2: ADHD vs. HC classification on Peking | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC (%) | ACC (%) | F1 (%) | SEN (%) | SPE (%) | PRE (%) | AUC (%) | ACC (%) | F1 (%) | SEN (%) | SPE (%) | PRE (%) | ||
| NF-SVM | 53.83∗ | 51.43 | 54.05 | 60.61 | 43.24 | 48.78 | 55.38∗ | 55.92 | 47.06 | 47.06 | 62.24 | 47.06 | |
| GF-SVM | 50.57∗ | 50.71 | 54.30 | 62.12 | 40.54 | 48.24 | 52.87∗ | 53.88 | 44.88 | 45.10 | 60.14 | 44.66 | |
| PCC-XGB | 57.76∗ | 53.57 | 55.78 | 62.12 | 45.95 | 50.62 | 55.74∗ | 54.29 | 49.55 | 53.92 | 54.55 | 45.83 | |
| PCC-RF | 64.99∗ | 61.43 | 57.81 | 56.06 | 66.22 | 59.68 | 54.37∗ | 54.29 | 42.86 | 41.18 | 63.64 | 44.68 | |
| MaLRR | 66.57∗ | 62.14 | 61.31 | 63.64 | 60.81 | 59.15 | 58.82∗ | 57.55 | 49.02 | 49.02 | 63.64 | 49.02 | |
| DANN | 67.22±3.04∗ | 62.57±2.46 | 61.98±5.93 | 67.58±16.78 | 58.11±19.11 | 62.33±10.36 | 59.70±2.34∗ | 53.55±6.67 | 43.35±16.87 | 53.92±33.77 | 53.29±34.23 | 48.16±4.75 | |
| RAINCOAT | 65.11±3.37∗ | 58.71±3.21 | 56.08±11.02 | 60.61±20.53 | 57.03±19.37 | 57.07±4.41 | 54.92±2.42∗ | 53.88±2.18 | 48.85±2.53 | 53.14±5.66 | 54.41±6.48 | 45.50±1.82 | |
| UFA-Net | 54.36±2.10∗ | 54.71±1.84 | 65.95±2.33 | 52.44±2.49 | 56.10±1.48 | 34.05±2.33 | 53.24±1.66∗ | 52.24±2.93 | 47.05±8.12 | 53.73±16.48 | 51.19±15.70 | 44.01±1.74 | |
| SHOT | 67.84±3.90∗ | 55.57±5.39 | 53.99±18.92 | 67.58±31.62 | 44.86±33.43 | 57.35±9.76 | 56.23±0.77∗ | 54.12±1.64 | 50.45±0.96 | 56.08±0.96 | 52.73±2.95 | 45.88±1.48 | |
| CDCL | 68.10±1.13∗ | 63.00±1.88 | 61.67±4.69 | 64.85±13.53 | 61.35±14.34 | 61.20±4.75 | 58.22±1.53∗ | 53.63±6.29 | 45.15±16.03 | 55.88±31.48 | 52.03±32.12 | 47.55±6.98 | |
| SCDA-Naive | 66.70±4.06∗ | 62.71±5.26 | 63.34±4.60 | 68.48±8.10 | 57.57±11.10 | 59.63±6.05 | 58.04±2.17∗ | 53.88±5.97 | 45.95±13.02 | 54.90±28.70 | 53.15±29.97 | 48.86±7.10 | |
| SCDA (Ours) | 72.79±2.10 | 68.00±0.83 | 66.77±2.74 | 68.79±7.94 | 67.30±6.53 | 65.48±1.92 | 61.56±0.93 | 58.20±3.78 | 51.18±7.32 | 56.27±20.16 | 59.58±20.79 | 52.23±5.79 | |
IV-C Experimental Settings
Six evaluation metrics are used to validate the effectiveness of our SCDA, including the area under the receiver operating characteristic curve (AUC), classification accuracy (ACC), F1 score (F1), sensitivity (SEN), specificity (SPE), and precision (PRE). Denote true positive as the number of subjects that are correctly classified as the positive category (i.e., disease group), true negative as the number of subjects that are correctly classified as the negative category (i.e., HC), false positive as the number of subjects that are wrongly classified as the positive group, and false negative as the number of subjects that are wrongly classified as the negative category. Here, ACC = , F1 = , SEN = , SPE = , and PRE =.
IV-D Results of Cross-Scanner Prediction
| Method | Task 1: ASD vs. HC classification on NYU1 | Task 2: T2DM-CI vs. HC on T2DM | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC (%) | ACC (%) | F1 (%) | SEN (%) | SPE (%) | PRE (%) | AUC (%) | ACC (%) | F1 (%) | SEN (%) | SPE (%) | PRE (%) | ||
| NF-SVM | 51.04∗ | 54.86 | 47.68 | 48.00 | 60.00 | 47.37 | 55.16∗ | 54.10 | 57.58 | 63.33 | 45.16 | 52.78 | |
| GF-SVM | 51.67∗ | 55.43 | 56.18 | 66.67 | 47.00 | 48.54 | 54.52∗ | 52.46 | 56.72 | 63.33 | 41.94 | 51.35 | |
| PCC-XGB | 53.20∗ | 49.71 | 49.43 | 57.33 | 44.00 | 43.43 | 54.73∗ | 55.74 | 57.14 | 60.00 | 51.61 | 54.55 | |
| PCC-RF | 54.43∗ | 54.29 | 47.37 | 48.00 | 59.00 | 46.75 | 54.30∗ | 55.74 | 55.74 | 56.67 | 54.84 | 54.84 | |
| MaLRR | 71.69∗ | 66.86 | 63.75 | 68.00 | 66.00 | 60.00 | 55.11∗ | 52.46 | 56.72 | 63.33 | 41.94 | 51.35 | |
| DANN | 63.92±3.00∗ | 59.20±4.94 | 56.21±5.36 | 63.20±17.25 | 56.20±20.39 | 53.85±6.04 | 59.70±3.67∗ | 54.43±4.32 | 47.10±16.77 | 50.00±30.98 | 58.71±31.30 | 55.26±6.12 | |
| RAINCOAT | 54.60±3.90∗ | 51.77±3.04 | 50.02±4.36 | 57.60±12.38 | 47.40±13.92 | 45.53±2.44 | 54.67±5.58∗ | 55.08±3.68 | 57.35±8.94 | 64.67±18.45 | 45.81±13.75 | 53.46±2.74 | |
| UFA-Net | 56.18±3.36∗ | 55.09±1.06 | 62.80±4.26 | 47.48±1.27 | 60.29±0.80 | 37.20±4.26 | 56.17±2.55∗ | 52.79±3.34 | 34.19±6.64 | 51.36±2.34 | 56.27±6.21 | 65.81±6.64 | |
| SHOT | 72.78±0.50∗ | 65.49±2.99 | 63.76±1.82 | 71.20±8.71 | 61.20±11.07 | 58.71±4.37 | 75.38±0.98 | 62.30±4.28 | 55.60±22.62 | 62.67±34.15 | 61.94±27.78 | 71.34±16.52 | |
| CDCL | 72.89±0.66∗ | 65.60±2.62 | 61.58±5.41 | 66.93±17.23 | 64.60±16.70 | 60.95±6.81 | 74.75±1.62∗ | 64.59±1.67 | 63.31±9.26 | 66.67±19.66 | 62.58±16.27 | 65.28±6.06 | |
| SCDA-Naive | 65.48±2.02∗ | 59.09±4.19 | 56.62±6.27 | 65.60±19.93 | 54.20±21.58 | 54.07±6.38 | 62.06±4.19∗ | 56.07±5.81 | 52.83±8.04 | 53.33±19.66 | 58.71±27.93 | 60.33±11.41 | |
| SCDA (Ours) | 73.31±0.53 | 67.20±2.25 | 63.85±1.48 | 68.00±8.43 | 66.60±10.13 | 61.28±4.43 | 75.63±0.66 | 65.57±2.74 | 65.74±2.60 | 68.00±10.87 | 63.23±14.80 | 66.16±8.75 | |
The classification results of our SCDA and eleven competing methods in cross-scanner prediction tasks are reported in Table IV. We also perform a two-tailed paired -test on the results of SCDA and each competing method, and ‘*’ denotes that the performance of SCDA is statistically significantly different () from a specific method. From Table IV, we have the following interesting observations.
First, our SCDA outperforms the first four traditional models (i.e., NF-SVM, GF-SVM, PCC-XGB, PCC-RF) by a large margin. The possible reason is that these methods only consider handcrafted features derived from a stationary functional connectivity matrix, without taking advantage of spatiotemporal characteristics inherent in fMRI timeseries. Despite the MaLRR method can encode spatiotemporal features, its feature learning and class prediction are performed in a separate manner, which may degrade the learning performance. Second, the SCDA generally achieves better results compared with three deep learning-based domain adaptation approaches (i.e., RAINCOAT, UFA-Net, and DANN). Note that RAINCOAT is originally designed for timeseries data and UFA-Net is specifically designed for fMRI data. Compared with these two methods, our SCDA exploits fMRI timeseries from more perspectives, resulting in general and diverse fMRI representations, which may facilitate effective cross-domain knowledge transfer. Furthermore, the two source-free domain adaptation approaches (i.e., SHOT and CDCL) show inferior performance compared with our SCDA. The possible reason may be that SHOT is optimized with information maximization, which may assign a high confidence score to incorrect prediction, leading to suboptimal results. And the CDCL method generates source prototypical features from a pretrained source model to initialize category centers of target data, which may introduce bias when there exists a large domain gap. This bias could negatively affect the adaptation process, as the initial centers may not accurately represent the target domain. Compared with these two methods, our SCDA neither utilizes information maximization nor category center initialization. Instead, the SCDA introduces a multi-perspective feature enrichment method to exploit fMRI characteristics inherent in target data based on stored source knowledge, demonstrating its superior performance. Additionally, our SCDA shows better results compared with SCDA-Naive which does not use unsupervised pretraining. The reason may be that, with unsupervised pretraining, SCDA can learn a general fMRI feature encoder from large-scale and diverse fMRI databases. And features derived from this encoder are expected to be less affected by domain bias, helping address cross-domain shift issues and improve target inference.

IV-E Results of Cross-Study Prediction
The classification results of our SCDA and eleven competing methods in cross-study prediction tasks are shown in Table V. From Table V, we have a similar observation to those in Table IV, that is, the proposed SCDA produces the overall best performance in most cases. Besides, we find that the standard deviation of our SCDA is smaller than that of the most competing methods, which indicates that the SCDA yields more stable and consistent classification results under different parameter initialization, suggesting the robustness of our method. In addition, we observe that SCDA-Naive has comparative results with DANN which has access to labeled source domain and unlabeled target domain. Note that in our work, we try to make SCDA-Naive and DANN as comparable as possible. Specifically, both of them extract fMRI features from multiple perspectives (i.e., window warping, receptive field manipulation, and window slicing), and they are both trained from scratch without the unsupervised pretraining strategy. Under such circumstances, our SCDA-Naive trained only based on a pretrained source model and unlabeled target data can achieve competitive results with DANN, which further demonstrates its effectiveness. Futhermore, our SCDA generally produces promising classification performance in cross-study prediction tasks, where the target domain is usually with a limited sample size (e.g., 61 in T2DM). This shows that our method can effectively improve inference of a small target dataset only based on a pretrained source model, which helps preserve privacy concerns and save storage burdens.
V Discussion
V-A Ablation Study
We conduct ablation studies to investigate the influence of several key components of the proposed SCDA, by comparing SCDA with its six variants called SCDAw/oDA, SCDAw/oS, SCDAw/oT, SCDA-WR, SCDA-WS, and SCDA-RS. Specifically, SCDAw/oDA denotes the SCDA without any adaptation technique, that is, the model trained on the source domain is directly applied to the target domain. SCDAw/oS means SCDA without spatial attention across ROIs, and SCDAw/oT means SCDA without temporal attention across different time windows. SCDA-WR, SCDA-WS, and SCDA-RS represent that SCDA extracts fMRI features from any pair of perspectives, i.e., window warping (W) and receptive field manipulation (R), window warping (W) and window slicing (S), and receptive field manipulation (R) and window slicing (S), respectively. Taking SCDA-WR for instance, we construct a two-branch feature enrichment architecture and use fMRI features with window warping (W) and receptive field manipulation (R) as inputs. Also, a mutual-consistency constraint is leveraged to reduce feature distribution gap across these two branches.
The average results of the SCDA and its variants in both cross-scanner and cross-study prediction tasks are reported in Fig. 3. For cross-scanner prediction, NYU1 and UM are used as source and target domains, respectively, and the task is to differentiate ASD patients from HCs in UM. For cross-study prediction, NYU2 is used as source domain and T2DM is used as target domain, and the task is to identify T2DM-CI patients from HCs in T2DM. From Fig. 3, we can see that SCDA is superior to SCDAw/oDA, and the underlying reason is that our SCDA considers cross-domain data heterogeneity by leveraging fMRI features inherent in the target domain, which helps improve target inference. Besides, the performance of SCDA is better than SCDAw/oS and SCDAw/oT. This implies that using spatial and temporal attention improves diagnostic performance in SCDA, which can help the model focus on discriminative brain regions and time windows for prediction. Furthermore, we can observe that SCDA consistently outperforms its degraded variants that capture fMRI representations from only two perspectives (i.e., SCDA-WR, SCDA-WS, and SCDA-RS). This indicates that our proposed multi-perspective feature enrichment strategy could learn complementary features from different views, and the collaboration of these features can greatly improve cross-domain prediction.
V-B Effect of Involved Databases in Unsupervised Pretraining
| Method | AUC (%) | ACC (%) | F1 (%) | SEN (%) | SPE (%) | PRE (%) |
|---|---|---|---|---|---|---|
| SCDA-ABIDE∗ | 70.50±1.27 | 65.14±3.54 | 64.47±2.62 | 67.58±10.08 | 62.97±14.42 | 63.11±5.27 |
| SCDA-MDD∗ | 71.02±1.65 | 64.86±2.41 | 62.82±6.03 | 65.45±16.33 | 64.32±16.03 | 63.67±5.32 |
| SCDA-ADHD∗ | 69.18±2.49 | 64.57±2.98 | 64.17±2.09 | 67.27±4.13 | 62.16±7.40 | 61.66±3.67 |
| SCDA | 72.79±2.10 | 68.00±0.83 | 66.77±2.74 | 68.79±7.94 | 67.30±6.53 | 65.48±1.92 |
As mentioned in Section III-C, we propose an unsupervised pretraining strategy based on 3,806 subjects from three large-scale auxiliary fMRI databases (i.e., ABIDE, REST-meta-MDD, and ADHD-200) to facilitate more comprehensive fMRI feature extraction. We now investigate the impact of involved databases in differentiating ASD patients from HCs in cross-scanner prediction tasks, with results shown in Table VI. The SCDA-ABIDE, SCDA-MDD, and SCDA-ADHD denote that only one specific database participates in unsupervised pretraining. Except for databases used in pretraining, all other settings of these methods remain the same.
As shown in Table VI, the SCDA pretrained with three databases generally outperforms its variants with one database involved for unsupervised pretraining. This implies that data scale and data diversity play important roles in pretraining a general fMRI feature encoder to improve cross-domain prediction. It’s interesting to explore more fMRI databases (e.g., Human Connectome Project444https://www.humanconnectome.org/) to further increase data size and enhance data diversity, which will be our future work.
V-C Influence of Source Model Initialization
| Method | AUC (%) | ACC (%) | F1 (%) | SEN (%) | SPE (%) | PRE (%) |
|---|---|---|---|---|---|---|
| SCDA-W∗ | 71.37±0.85 | 65.43±2.10 | 63.57±4.38 | 65.15±11.46 | 65.68±10.93 | 63.54±3.72 |
| SCDA-R∗ | 67.10±1.25 | 61.86±2.66 | 59.87±7.98 | 63.64±17.17 | 60.27±17.61 | 61.02±6.58 |
| SCDA-S∗ | 69.25±2.10 | 63.29±4.18 | 61.49±5.16 | 63.64±13.99 | 62.97±17.69 | 62.22±6.08 |
| SCDA | 72.79±2.10 | 68.00±0.83 | 66.77±2.74 | 68.79±7.94 | 67.30±6.53 | 65.48±1.92 |

In this work, we first construct an initial MFE via unsupervised pretraining based on three auxiliary fMRI databases. Then, we initialize the source model using average parameters of three branches of MFE. In this section, to examine the influence of different initialized source models on classification performance, we use parameters of each branch of MFE to initialize the source model rather than the average of three branches. The results of our method and its three variants (called SCDA-W, SCDA-R, SCDA-S) in ASD vs. HC classification in the cross-scanner prediction task are shown in Table VII. Here, SCDA-W, SCDA-R, and SCDA-S denote that we initialize the source model using the branch with window warping (W), receptive field manipulation (R), and window slicing (S) for fMRI feature representation learning, respectively. From Table VII, we can see that our SCDA consistently outperforms these competing methods. This result is reasonable since averaged parameters from multiple branches can help extract more general fMRI features, which can facilitate more effective cross-domain knowledge transfer.
V-D Visualization of fMRI Features
We employ t-SNE [43] to visualize fMRI feature distributions before and after domain adaptation (via SCDA) in cross-scanner and cross-study prediction tasks, with results shown in Fig. 4. In the cross-scanner prediction task, NYU1 and UM are used as source and target domains, respectively. In the cross-study prediction task, Site-21 and NYU1 serve as source and target domains, respectively. In our study, each subject’s fMRI timeseries is represented by an matrix, with time points and brain ROIs. Before domain adaptation, we represent each subject using the flattened vector from the matrix. Then, we optimize SCDA using unlabeled target data and finally derive a feature encoder. We use this feature encoder to capture fMRI feature representation, which is used to represent each subject after domain adaptation. In Fig. 4, different colors represent different domains, i.e., magenta and blue denote source and target domains, respectively. It can be seen from Fig. 4 that after adaptation using our SCDA, features learned from source and target domains are mixed and mapped into a common latent feature space. It implies that our SCDA helps reduce the cross-domain data distribution gap.
V-E Identified Discriminative Brain ROIs

We employ the squeeze-excitation mechanism [32] to automatically learn spatial attention across different brain ROIs (=116), with each element denoting the discriminative ability of the corresponding brain ROI for prediction. We visualize the top 10 important brain ROIs identified by our SCDA in two cross-scanner prediction tasks in Fig. 5 (a) and Fig. 5 (b), respectively. The first task is ASD diagnosis on the UM site and the second task is ADHD diagnosis on the Peking site. We show the averaged importance score of correctly-classified subjects with a disease, where a larger circle denotes more important ROIs, corresponding to a higher value.
From Fig. 5 (a), we can see that right paracentral lobule (PCL.R) plays the most important role in identifying ASD patients from HCs. This finding is consistent with a previous study [44], where this brain region is reported to exhibit consistently lower resting-state local connectivity in ASD subjects compared to typically developing controls. The reason may be that the paracentral lobule can operate with precuneus to produce a sense of self in a spatial world. Its impairment may disrupt the body and spatial representation in ASD patients and affect their development of theory of mind [45]. Moreover, functional abnormalities of our identified ROIs, e.g., middle frontal gyrus, orbital part (ORBmid.L), temporal pole: middle temporal gyrus (TPOmid.R), inferior parietal, supramarginal and angular gyri (IPL.L), middle frontal gyrus (MFG.R), have been verified in previous studies [46, 47, 48, 49]. These regions may be used as potential biomarkers to improve ASD diagnosis.
From Fig. 5 (b), we find that anterior cingulate and paracingulate gyri (ACG.L) plays the most important role in ADHD diagnosis, which is consistent with [50]. The possible reason may be that ACG involves various brain functions, e.g., executive control and cognitive processing, and its abnormality may lead to the breakdown of attention allocation and cognition modulation, causing inattention and hyperactivity in ADHD patients [50]. Besides, other identified ROIs by our SCDA, e.g., insula (INS.L), lenticular nucleus, pallidum (PAL.L), superior frontal gyrus, medial (SFGmed.L), have also been verified associated with functional abnormalities in previous ADHD studies [51, 52, 53].
V-F Limitations and Future Work
There are several limitations in the current study. First, our SCDA is a white-box source-free adaptation approach, where the parameters of the source model are accessible, which may suffer from data leakage problems under membership inference attack [54]. In the future, we will investigate source-free adaptation in black-box settings, where only prediction of the source model is available, which could further help protect data privacy in practical scenarios. Second, this work explores three feature enrichment strategies to exploit fMRI representations for target model construction. We will investigate more feature enrichment techniques (e.g., magnitude warping [55]) to further facilitate fMRI learning and enhance target inference. In addition, our SCDA only leverages functional MRI for disease diagnosis without taking advantage of structural MRI data and/or non-imaging information (e.g., age, gender). Future work will seek to combine complementary feature characteristics from different modalities to further improve learning performance. Lastly, current work learns brain functional connectivity features based on a single brain atlas (i.e., AAL) with 116 brain ROIs. In the future, we will incorporate fMRI representations from multi-scale atlases for brain parcellation, which may provide different fine-grained information to further boost classification performance.
VI Conclusions
This paper introduces a source-free collaborative domain adaptation (SCDA) framework for fMRI-based neurological disorder diagnosis. Our SCDA achieves source-to-target domain adaptation only based on a pretrained source model and unlabeled target data. Specifically, a multi-perspective feature enrichment method is designed, which contains multiple collaborative branches to dynamically exploit target data from different views. The parameters of MFE initialized by the pretrained source model can be fine-tuned on unlabeled target fMRI, thus catering to data distributions of the target domain. Furthermore, we propose an unsupervised pretraining strategy to leverage large-scale auxiliary fMRIs from three public databases to further improve fMRI representation learning. Extensive experiments in cross-scanner and cross-study prediction tasks show the effectiveness of our proposed method.
References
- [1] C. Lord, T. S. Brugha, T. Charman, J. Cusack, G. Dumas, T. Frazier, E. J. Jones, R. M. Jones, A. Pickles, M. W. State et al., “Autism spectrum disorder,” Nature Reviews Disease Primers, vol. 6, no. 1, pp. 1–23, 2020.
- [2] W. M. Van Der Flier, I. Skoog, J. A. Schneider, L. Pantoni, V. Mok, C. L. Chen, and P. Scheltens, “Vascular cognitive impairment,” Nature Reviews Disease Primers, vol. 4, no. 1, pp. 1–16, 2018.
- [3] M. D. Fox and M. E. Raichle, “Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging,” Nature Reviews Neuroscience, vol. 8, no. 9, pp. 700–711, 2007.
- [4] G. Deshpande, P. Wang, D. Rangaprakash, and B. Wilamowski, “Fully connected cascade artificial neural network architecture for attention deficit hyperactivity disorder classification from functional magnetic resonance imaging data,” IEEE Transactions on Cybernetics, vol. 45, no. 12, pp. 2668–2679, 2015.
- [5] S. Gallo, A. ElGazzar, P. Zhutovsky, R. M. Thomas, N. Javaheripour, L. Meng, L. Bartova, D. R. Bathula, U. Dannlowski, C. Davey et al., “Thalamic hyperconnectivity as neurophysiological signature of major depressive disorder in two multicenter studies,” PsyArXiv, 2021, doi:10.31234/osf.io/b6a7q.
- [6] Y. Shi, L. Zhang, Z. Wang, X. Lu, T. Wang, D. Zhou, and Z. Zhang, “Multivariate machine learning analyses in identification of major depressive disorder using resting-state functional connectivity: A multicentral study,” ACS Chemical Neuroscience, vol. 12, no. 15, pp. 2878–2886, 2021.
- [7] A. Yamashita, N. Yahata, T. Itahashi, G. Lisi, T. Yamada, N. Ichikawa, M. Takamura, Y. Yoshihara, A. Kunimatsu, N. Okada et al., “Harmonization of resting-state functional MRI data across multiple imaging sites via the separation of site differences into sampling bias and measurement bias,” PLoS Biology, vol. 17, no. 4, p. e3000042, 2019.
- [8] Y.-L. Huang, W.-T. Hsieh, H.-C. Yang, and C.-C. Lee, “Conditional domain adversarial transfer for robust cross-site ADHD classification using functional MRI,” in ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE, 2020, pp. 1190–1194.
- [9] Y. Gao, Y. Zhang, Z. Cao, X. Guo, and J. Zhang, “Decoding brain states from fMRI signals by using unsupervised domain adaptation,” IEEE Journal of Biomedical and Health Informatics, vol. 24, no. 6, pp. 1677–1685, 2019.
- [10] M. Bateson, H. Kervadec, J. Dolz, H. Lombaert, and I. B. Ayed, “Source-free domain adaptation for image segmentation,” Medical Image Analysis, vol. 82, p. 102617, 2022.
- [11] H. Xia, H. Zhao, and Z. Ding, “Adaptive adversarial network for source-free domain adaptation,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 2021, pp. 9010–9019.
- [12] C. Craddock, Y. Benhajali, C. Chu, F. Chouinard, A. Evans, A. Jakab, B. S. Khundrakpam, J. D. Lewis, Q. Li, M. Milham et al., “The neuro bureau preprocessing initiative: Open sharing of preprocessed neuroimaging data and derivatives,” Frontiers in Neuroinformatics, vol. 7, p. 27, 2013.
- [13] C.-G. Yan, X. Chen, L. Li, F. X. Castellanos, T.-J. Bai, Q.-J. Bo, J. Cao, G.-M. Chen, N.-X. Chen, W. Chen et al., “Reduced default mode network functional connectivity in patients with recurrent major depressive disorder,” Proceedings of the National Academy of Sciences, vol. 116, no. 18, pp. 9078–9083, 2019.
- [14] P. Bellec, C. Chu, F. Chouinard-Decorte, Y. Benhajali, D. S. Margulies, and R. C. Craddock, “The neuro bureau ADHD-200 preprocessed repository,” NeuroImage, vol. 144, pp. 275–286, 2017.
- [15] M. Zhao, W. Yan, N. Luo, D. Zhi, Z. Fu, Y. Du, S. Yu, T. Jiang, V. D. Calhoun, and J. Sui, “An attention-based hybrid deep learning framework integrating brain connectivity and activity of resting-state functional MRI data,” Medical Image Analysis, vol. 78, p. 102413, 2022.
- [16] H. Cui, W. Dai, Y. Zhu, X. Kan, A. A. C. Gu, J. Lukemire, L. Zhan, L. He, Y. Guo, and C. Yang, “BrainGB: A benchmark for brain network analysis with graph neural networks,” IEEE Transactions on Medical Imaging, vol. 42, pp. 493–506, 2022.
- [17] M. Wang, D. Zhang, J. Huang, P.-T. Yap, D. Shen, and M. Liu, “Identifying autism spectrum disorder with multi-site fMRI via low-rank domain adaptation,” IEEE Transactions on Medical Imaging, vol. 39, no. 3, pp. 644–655, 2019.
- [18] J. Zhang, P. Wan, and D. Zhang, “Transport-based joint distribution alignment for multi-site autism spectrum disorder diagnosis using resting-state fMRI,” in International Conference on Medical Image Computing and Computer Assisted Intervention. Springer, 2020, pp. 444–453.
- [19] Y. Fang, M. Wang, G. G. Potter, and M. Liu, “Unsupervised cross-domain functional MRI adaptation for automated major depressive disorder identification,” Medical Image Analysis, vol. 84, p. 102707, 2023.
- [20] T. Zhang, Z. Zhao, C. Zhang, J. Zhang, Z. Jin, and L. Li, “Classification of early and late mild cognitive impairment using functional brain network of resting-state fMRI,” Frontiers in Psychiatry, vol. 10, p. 572, 2019.
- [21] L. Torlay, M. Perrone-Bertolotti, E. Thomas, and M. Baciu, “Machine learning-XGBoost analysis of language networks to classify patients with epilepsy,” Brain Informatics, vol. 4, no. 3, pp. 159–169, 2017.
- [22] P. K. Douglas, S. Harris, A. Yuille, and M. S. Cohen, “Performance comparison of machine learning algorithms and number of independent components used in fMRI decoding of belief vs. disbelief,” NeuroImage, vol. 56, no. 2, pp. 544–553, 2011.
- [23] H. Zhang, R. Song, L. Wang, L. Zhang, D. Wang, C. Wang, and W. Zhang, “Classification of brain disorders in rs-fMRI via local-to-global graph neural networks,” IEEE Transactions on Medical Imaging, vol. 42, pp. 444–455, 2023.
- [24] X. Li, Y. Zhou, N. Dvornek, M. Zhang, S. Gao, J. Zhuang, D. Scheinost, L. H. Staib, P. Ventola, and J. S. Duncan, “BrainGNN: Interpretable brain graph neural network for fMRI analysis,” Medical Image Analysis, vol. 74, p. 102233, 2021.
- [25] B.-H. Kim, J. C. Ye, and J.-J. Kim, “Learning dynamic graph representation of brain connectome with spatio-temporal attention,” Advances in Neural Information Processing Systems, vol. 34, pp. 4314–4327, 2021.
- [26] Y. Zhang, H. Zhang, E. Adeli, X. Chen, M. Liu, and D. Shen, “Multiview feature learning with multiatlas-based functional connectivity networks for MCI diagnosis,” IEEE Transactions on Cybernetics, vol. 52, no. 7, pp. 6822–6833, 2020.
- [27] T. Wang, A. Bezerianos, A. Cichocki, and J. Li, “Multikernel capsule network for schizophrenia identification,” IEEE Transactions on Cybernetics, vol. 52, no. 6, pp. 4741–4750, 2020.
- [28] R. Liu, Z.-A. Huang, Y. Hu, Z. Zhu, K.-C. Wong, and K. C. Tan, “Spatial-temporal co-attention learning for diagnosis of mental disorders from resting-state fMRI data,” IEEE Transactions on Neural Networks and Learning Systems, 2023.
- [29] Z. Luo, G. Chen, Y. Jia, S. Zhong, J. Gong, F. Chen, J. Wang, Z. Qi, X. Liu, L. Huang et al., “Shared and specific dynamics of brain segregation and integration in bipolar disorder and major depressive disorder: A resting-state functional magnetic resonance imaging study,” Journal of Affective Disorders, vol. 280, pp. 279–286, 2021.
- [30] J. Bai, W. Yu, Z. Xiao, V. Havyarimana, A. C. Regan, H. Jiang, and L. Jiao, “Two-stream spatial-temporal graph convolutional networks for driver drowsiness detection,” IEEE Transactions on Cybernetics, vol. 52, no. 12, pp. 13 821–13 833, 2021.
- [31] K. Xu, W. Hu, J. Leskovec, and S. Jegelka, “How powerful are graph neural networks?” arXiv preprint arXiv:1810.00826, 2018.
- [32] J. Hu, L. Shen, and G. Sun, “Squeeze-and-excitation networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018, pp. 7132–7141.
- [33] S. Ioffe and C. Szegedy, “Batch normalization: Accelerating deep network training by reducing internal covariate shift,” in International Conference on Machine Learning. PMLR, 2015, pp. 448–456.
- [34] D. Hendrycks and K. Gimpel, “Gaussian error linear units (GELUs),” arXiv preprint arXiv:1606.08415, 2016.
- [35] A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, L. Kaiser, and I. Polosukhin, “Attention is all you need,” Advances in Neural Information Processing Systems, vol. 30, 2017.
- [36] A. Le Guennec, S. Malinowski, and R. Tavenard, “Data augmentation for time series classification using convolutional neural networks,” in ECML/PKDD Workshop on Advanced Analytics and Learning on Temporal Data, 2016.
- [37] E. O. Brigham, The fast Fourier transform and its applications. Prentice-Hall, Inc., 1988.
- [38] Y. Ganin, E. Ustinova, H. Ajakan, P. Germain, H. Larochelle, F. Laviolette, M. Marchand, and V. Lempitsky, “Domain-adversarial training of neural networks,” The Journal of Machine Learning Research, vol. 17, no. 1, pp. 2096–2030, 2016.
- [39] H. He, O. Queen, T. Koker, C. Cuevas, T. Tsiligkaridis, and M. Zitnik, “Domain adaptation for time series under feature and label shifts,” in International Conference on Machine Learning. PMLR, 2023.
- [40] J. Liang, D. Hu, and J. Feng, “Do we really need to access the source data? Source hypothesis transfer for unsupervised domain adaptation,” in International Conference on Machine Learning. PMLR, 2020, pp. 6028–6039.
- [41] R. Wang, Z. Wu, Z. Weng, J. Chen, G.-J. Qi, and Y.-G. Jiang, “Cross-domain contrastive learning for unsupervised domain adaptation,” IEEE Transactions on Multimedia, pp. 1665–1673, 2022.
- [42] T. Chen and C. Guestrin, “XGBoost: A scalable tree boosting system,” in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, pp. 785–794.
- [43] L. Van der Maaten and G. Hinton, “Visualizing data using t-SNE,” Journal of Machine Learning Research, vol. 9, no. 11, pp. 2579–2605, 2008.
- [44] L. E. Libero, M. Schaer, D. D. Li, D. G. Amaral, and C. W. Nordahl, “A longitudinal study of local gyrification index in young boys with autism spectrum disorder,” Cerebral Cortex, vol. 29, no. 6, pp. 2575–2587, 2019.
- [45] W. Cheng, E. T. Rolls, H. Gu, J. Zhang, and J. Feng, “Autism: Reduced connectivity between cortical areas involved in face expression, theory of mind, and the sense of self,” Brain, vol. 138, no. 5, pp. 1382–1393, 2015.
- [46] S. Xu, M. Li, C. Yang, X. Fang, M. Ye, L. Wei, J. Liu, B. Li, Y. Gan, B. Yang et al., “Altered functional connectivity in children with low-function autism spectrum disorders,” Frontiers in Neuroscience, vol. 13, p. 806, 2019.
- [47] J. Xu, C. Wang, Z. Xu, T. Li, F. Chen, K. Chen, J. Gao, J. Wang, and Q. Hu, “Specific functional connectivity patterns of middle temporal gyrus subregions in children and adults with autism spectrum disorder,” Autism Research, vol. 13, no. 3, pp. 410–422, 2020.
- [48] N. F. Wymbs, M. B. Nebel, J. B. Ewen, and S. H. Mostofsky, “Altered inferior parietal functional connectivity is correlated with praxis and social skill performance in children with autism spectrum disorder,” Cerebral Cortex, vol. 31, no. 5, pp. 2639–2652, 2021.
- [49] A. Crider, R. Thakkar, A. O. Ahmed, and A. Pillai, “Dysregulation of estrogen receptor beta (ER), aromatase (CYP19A1), and ER co-activators in the middle frontal gyrus of autism spectrum disorder subjects,” Molecular Autism, vol. 5, no. 1, pp. 1–10, 2014.
- [50] N. Makris, L. J. Seidman, E. M. Valera, J. Biederman, M. C. Monuteaux, D. N. Kennedy, V. S. Caviness Jr, G. Bush, K. Crum, A. B. Brown et al., “Anterior cingulate volumetric alterations in treatment-naive adults with ADHD: A pilot study,” Journal of Attention Disorders, vol. 13, no. 4, pp. 407–413, 2010.
- [51] X. Gao, M. Zhang, Z. Yang, M. Wen, H. Huang, R. Zheng, W. Wang, Y. Wei, J. Cheng, S. Han et al., “Structural and functional brain abnormalities in internet gaming disorder and attention-deficit/hyperactivity disorder: A comparative meta-analysis,” Frontiers in Psychiatry, vol. 12, p. 679437, 2021.
- [52] F. Samea, S. Soluki, V. Nejati, M. Zarei, S. Cortese, S. B. Eickhoff, M. Tahmasian, and C. R. Eickhoff, “Brain alterations in children/adolescents with ADHD revisited: A neuroimaging meta-analysis of 96 structural and functional studies,” Neuroscience & Biobehavioral Reviews, vol. 100, pp. 1–8, 2019.
- [53] K. Jiang, Y. Yi, L. Li, H. Li, H. Shen, F. Zhao, Y. Xu, and A. Zheng, “Functional network connectivity changes in children with attention-deficit hyperactivity disorder: A resting-state fMRI study,” International Journal of Developmental Neuroscience, vol. 78, pp. 1–6, 2019.
- [54] M. Nasr, R. Shokri, and A. Houmansadr, “Comprehensive privacy analysis of deep learning: Passive and active white-box inference attacks against centralized and federated learning,” in 2019 IEEE Symposium on Security and Privacy (SP). IEEE, 2019, pp. 739–753.
- [55] T. T. Um, F. M. Pfister, D. Pichler, S. Endo, M. Lang, S. Hirche, U. Fietzek, and D. Kulić, “Data augmentation of wearable sensor data for Parkinson’s disease monitoring using convolutional neural networks,” in Proceedings of the 19th ACM International Conference on Multimodal Interaction, 2017, pp. 216–220.