RespireNet: A Deep Neural Network for Accurately Detecting Abnormal Lung Sounds in Limited Data Setting
Abstract
Auscultation of respiratory sounds is the primary tool for screening and diagnosing lung diseases. Automated analysis, coupled with digital stethoscopes, can play a crucial role in enabling tele-screening of fatal lung diseases. Deep neural networks (DNNs) have shown a lot of promise for such problems, and are an obvious choice. However, DNNs are extremely data hungry, and the largest respiratory dataset ICBHI [21] has only 6898 breathing cycles, which is still small for training a satisfactory DNN model. In this work, RespireNet, we propose a simple CNN-based model, along with a suite of novel techniques—device specific fine-tuning, concatenation-based augmentation, blank region clipping, and smart padding—enabling us to efficiently use the small-sized dataset. We perform extensive evaluation on the ICBHI dataset, and improve upon the state-of-the-art results for 4-class classification by 2.2%.
Index Terms— Abnormality detection, lung sounds, crackle and wheeze, ICBHI dataset, deep learning
1 Introduction
Respiratory diseases like asthma, chronic obstructive pulmonary disease (COPD), lower respiratory tract infection, lung cancer, and tuberculosis are the leading causes of death worldwide [14], constituting four of the 12 most common causes of death. Early diagnosis has been found to be crucial in limiting the spread of respiratory diseases, and their adverse effects on the length and quality of life. Listening to chest sounds using a stethoscope is a standard method for screening and diagnosing lung diseases. It provides a low cost and non-invasive screening methodology, avoiding the exposure risks of radiography and patient-compliance requirements associated with tests such as Spirometry.
There are a few drawbacks of stethoscope-based diagnosis: requirement of a trained medical professional to interpret auscultation signals, and subjectivity in interpretations causing inter-listener variability. These limitations are exacerbated in impoverished settings and during pandemic situations (such as COVID-19), due to shortage of expert medical professionals. Automated analysis of respiratory sounds can alleviate these drawbacks, and also help in enabling tele-medicine applications to monitor patients outside a clinic by less-skilled workforce such as community health workers.

Algorithmic detection of lung diseases from respiratory sounds has been an active area of research [17, 20] especially with the advent of digital stethoscopes. Most of these works focus on detecting abnormal respiratory sounds of wheeze and crackle. Wheeze is a typical symptom of asthma and COPD. It is characterized by a high-pitched continuous sound in the frequency range of 100-2500Hz and duration above 80 msec [3, 19]. Crackles, which are associated with COPD, chronic bronchitis, pneumonia and lung fibrosis [7, 18], have a discontinuous, non-tonal sound, around frequency of 650 Hz and duration of 5 msec (for fine crackles), or frequency of 100-500 Hz and duration of 15 msec (for coarse crackles).
Although early works focused on hand-crafted features and traditional machine learning [8, 4], more recently, deep learning based methods have received the most attention [9, 1, 12]. For training DNNs, a time-frequency representation of the audio signal such as Mel-spectrograms [25, 10, 1], stacked MFCC features [25, 2, 16, 13, 9] or optimized S-transform spectrogram [6] is used. This 2D “image” is then fed into CNNs [16, 2], RNNs [15, 9], or hybrid CNN-RNNs [1] to learn robust high dimensional representations.
It is well known that DNNs are data hungry and typically require large datasets to achieve good performance. In this work, we use the ICBHI challenge dataset [21], a popular respiratory sound dataset. In spite of being the largest publicly available dataset, it has only 6898 breathing cycle samples, which is quite small for training deep networks. Thus, a big focus of our work has been on developing a suite of techniques to help train DNNs in a data efficient manner. We found that a simple CNN architecture, such as ResNet, is adequate for achieving good accuracy. This is in contrast to prior work employing complex architectures like hybrid CNN-RNN [1], non-local block additions to CNNs [12], etc.
In order to efficiently use the available data, we did extensive analysis of the ICBHI dataset. We found several characteristics of the data that might inhibit training DNNs effectively. For example, the dataset contains audio recordings from four different devices, with skewed distribution of samples across the devices, which makes it difficult for DNNs to generalize well across devices. Similarly, the dataset has a skewed distribution across normal and abnormal classes, and varying lengths of audio samples. We propose multiple novel techniques to address these problems—device specific fine-tuning, concatenation-based augmentation, blank region clipping, and smart padding. We perform extensive evaluation and ablation analysis of these techniques.
The main contributions of our work are:
-
1.
demonstration that a simple network architecture is sufficient for respiratory sound classification, and more focus is needed on making efficient use of available data.
-
2.
a detailed analysis of the ICBHI dataset pointing out its characteristics impacting DNN training significantly.
-
3.
a suite of techniques—device specific fine-tuning, concatenation-based augmentation, blank region clipping and smart padding—enabling efficient dataset usage. These techniques are orthogonal to the choice of network architecture and should be easy to incorporate in other networks.
2 Method
Dataset: We perform all evaluations on the ICBHI scientific challenge respiratory sound dataset [21, 22]. It is one of the largest publicly available respiratory datasets. The dataset comprises of 920 recordings from 126 patients with a combined total duration of 5.5 hours. Each breathing cycle in a recording is annotated by an expert as one of the four classes: normal, crackle, wheeze, or both (crackle and wheeze). The dataset comprises of recordings from four different devices111The four devices used for recordings are AKGC417L Microphone, 3M Littmann Classic II SE Stethoscope, 3M Litmmann 3200 Electronic Stethoscope, and WelchAllyn Meditron Master Elite Electronic Stethoscope from hospitals in Portugal and Greece. For every patient, data was recorded at seven different body locations.
Pre-processing: The sampling rate of recordings in the dataset varies from 4 kHz to 44.1 kHz. To standardize, we down-sample the recordings to 4 kHz and apply a 5-th order Butterworth band-pass filter to remove noise (heartbeat, background speech, etc.). We also apply standard normalization on the input signal to map the values within the range (-1, 1). The audio signal is then converted into a Mel-spectrogram, which is fed into our DNN.
Network architecture: We use a CNN-based network, ResNet-34, followed by two 128-d fully connected linear layers with ReLU activations. The last layer applies softmax activation to model classwise probabilities. Dropout is added to the fully-connected layers to prevent overfitting. The network is trained via a standard categorical cross-entropy loss to minimize the loss for multi-class classification. The overall framework and architecture is illustrated in Figure 1.
2.1 Efficient Dataset Utilization
Even though ICBHI is the largest publicly available dataset with 6898 samples, it is still relatively small for training DNNs effectively. Thus, a major focus of our work has been to develop techniques to efficiently use the available samples. We extensively analyzed the dataset to identify dataset characteristics that inhibit training DNNs effectively, and propose solutions to overcome the same.
The first commonly used technique we apply is transfer learning, where we initialize our network with weights of a pre-trained ResNet-34 network on ImageNet [23]. This is followed by our training where we train the entire network end-to-end. Interestingly, even though ImageNet dataset is very different from the spectrograms which our network sees, we still found this initialization to help significantly. Most likely, low level features such as edge-detection are still similar and thus “transfer” well.
Concatenation-based Augmentation: Like most medical datasets, ICBHI dataset has a huge class imbalance, with the normal class accounting for 53% of the samples. To prevent the model from overfitting on abnormal classes, we experimented with several data augmentation techniques. We first apply standard audio augmentation techniques, such as noise addition, speed variation, random shifting, pitch shift, etc., and also use a weighted random sampler to sample mini-batches uniformly from each class. These standard techniques help a little, but to further improve generalization of the underrepresented classes (wheeze, crackle, both), we developed a concatenation based augmentation technique where we generate a new sample of a class by randomly sampling two samples of the same class and concatenating them (see Figure 2). This scheme led to a non-trivial improvement in the classification accuracy of abnormal classes.

Smart Padding: The breathing cycle length varies across patients as well as within a patient depending on various factors (e.g., breathing rate can increase moderately during fever). In the ICBHI dataset, the length of breathing cycles ranges from 0.2s to 16.2s with a mean cycle length of 2.7s. This poses a problem while training our network as it expects a fixed size input222CNNs can be made size agnostic by using adaptive average pooling, but that typically hurts accuracy.. The standard way to handle this is to pad the audio signal to a fixed size via zero-padding or reflection based padding. We propose a novel smart padding scheme, which uses a variant of the augmentation scheme described above. For each data sample, smart padding first looks at the breathing cycle sample for the same patient taken just before and after the current one. If this neighbouring cycle is of the same class or of the normal class, we concatenate the current sample with it. If not, we pad by copying the same cycle again. We continue this process until we reach our desired size. This smart padding scheme also augments the data and helps prevent overfitting. We experimented with different input lengths, and found a 7s window to perform best. A small window led to clipping of samples, thus loosing valuable information in an already scarce dataset, while a very large window caused repetition leading to degraded performance.

Blank Region Clipping: On analyzing samples using Grad-Cam++ [5] which our base model mis-classified, we found notable black regions333Black region in a spectrogram means that the audio signal has zero energy in the corresponding audio frequency range. at higher frequency regions of their spectrograms (Figure 3). On further analysis, we found that many samples, and in particular 100% of the Litt3200 device samples, had blank region in the 1500-2000Hz frequency range. Since this was adversely affecting our network performance, we selectively clip off the blank rows from the high frequency regions of such spectrograms. This ensures that the network focuses on the region of interest leading to improved performance. Figure 3 shows this in action.
Device Specific Fine-tuning: The ICBHI dataset has samples from 4 different devices. We found that the distribution of samples across devices is heavily skewed, e.g. the AKGC417L Microphone alone contributes to 63% of the samples. Since each device has different audio characteristics, the DNN may fail to generalize across devices, especially for the underrepresented devices in the already small dataset. To verify this, we divided the test set into 4 subsets depending on their device type, and compute the accuracy of abnormal class samples in each subset. As expected, we found the classification accuracy to be strongly correlated with the training set size of the corresponding device. To address this, we first train a common model with the full training data (stage-1, Figure 1). We then make 4 copies of this model and fine-tune (stage-2) them for each device separately by using only the subset of training data for that device. We found this approach to significantly improve the performance, especially for the underrepresented devices.
3 Experiments
We evaluate the performance of our framework on the respiratory anomaly classification task proposed in the ICBHI challenge [21]. This is further divided into two subtasks: (i) classify a breathing cycle into one of the four classes–normal(N), crackle(C), wheeze(W), both(B), and (ii) classify a breathing cycle into normal or anomalous class, where . Our evaluation method is same as the one proposed in the original ICBHI challenge. The final score is computed as the mean of Sensitivity and Specificity , where and are the number of correctly classified and total number of samples in class , respectively (). For the 2-class case, we adopt the anomalous and normal class scores as and respectively, and the score is computed as their mean.
We compare our performance using the above evaluation metric on two dataset divisions: the official 60-40 split [21] and the 80-20 split [11, 12, 1] for train-test444For both the splits, the train and test set are patient-wise disjoint.. The Sensitivity , Specificity and ICBHI Score values are reported in Table 1. RespireNet achieves state-of-the-art (SOTA) in both train-test split divisions, and outperforms SOTA [12] on the official split (60-40) by and SOTA [1] on the 80-20 split by . Further, RespireNet achieves a score of on the 2-class classification task, achieving the new SOTA.
Implementation Details: We train our models on a Tesla v100 GPU on a Microsoft Azure VM. We used the SGD optimizer with momentum of 0.9, and a batch size of 64. We used a fixed learning rate of 1e-3 for stage-1 and 1e-4 for stage-2 of training. Stage-1 was trained for 200 epochs. The highest validation checkpoint from stage-1 was used to train stage-2 for another 50 epochs for each device.
| Split & Task | Method | Score | ||
|---|---|---|---|---|
| 6040 Split | Jakovljevic et al. [8] | - | - | 39.5% |
| & | Chambres et al. [4] | 78.1% | 20.8% | 49.4% |
| 4-class | Serbes et al. [24] | - | - | 49.9% |
| Ma et al. [11] | 69.2% | 31.1% | 50.2% | |
| Ma et al. [12] | 63.2% | 41.3% | 52.3% | |
| CNN (ours) | 71.4% | 39.0% | 55.2% | |
| CNN+CBA+BRC (ours) | 71.8% | 39.6% | 55.7% | |
| CNN+CBA+BRC+FT (ours) | 72.3% | 40.1% | 56.2% | |
| 8020 Split | Kochetov et al. [9] | 73.0% | 58.4% | 65.7 % |
| & | Acharya et al. [1] | 84.1% | 48.6% | 66.3% |
| 4-class | Ma et al. [12] | 64.7% | 63.7% | 64.2% |
| CNN (ours) | 78.8% | 53.6% | 66.2% | |
| CNN+CBA+BRC (ours) | 79.7% | 54.4% | 67.1% | |
| CNN+CBA+BRC+FT (ours) | 83.3% | 53.7% | 68.5% | |
| 8020 Split | CNN (ours) | 83.3% | 60.5% | 71.9% |
| & | CNN+CBA+BRC (ours) | 76.4% | 71.0% | 73.7% |
| 2-class | CNN+CBA+BRC+FT (ours) | 80.9% | 73.1% | 77.0% |
We further analyze the effect of our novel proposed techniques by conducting an ablation analysis on the 4-class classification task on the 80/20 split.
Concatenation-based Augmentation: Due to the small size of abnormal samples in the dataset, our model tends to overfit on the abnormal classes quickly, and achieved a score of . Standard augmentations (noise addition, etc.) improved the score to , which further improved to with our concatenation-based augmentation. Also, most of the gain came from improved accuracy of the abnormal classes, where the sensitivity increased by 1.5%. This demonstrates that our augmentation scheme to generate novel samples for the abnormal class helps the model generalize better.
| Length. | 1 sec | 2 sec | 3 sec | 4 sec | 5 sec | 6 sec | 7 sec | 8 sec | 9 sec |
|---|---|---|---|---|---|---|---|---|---|
| Scores | 56.6 | 59.0 | 60.3 | 61.1 | 62.3 | 64.4 | 66.2 | 65.1 | 65.5 |
Smart Padding: The length of breathing cycle in the dataset has a wide variation, thus we need to pad the shorter samples and clip the longer ones to match the input length of the network. We experimented with different input lengths and found that a 7s length performed optimally (see Table 2). Since the average cycle length is 2.7s, padding became crucial as a majority of the inputs need padding. We found the padding scheme to have a significant impact on accuracy. For the base model, smart padding improves accuracy over zero-padding and reflection-based padding by 5% and 2% respectively. This demonstrates the effectiveness of our padding scheme, which incorporates data augmentation for padding, rather than plain copying or adding zeros.
Blank Region Clipping: This provided an improvement of over the base model score of . When combined with our proposed augmentation, it helped achieve a score of , outperforming the current SOTA [1] by .
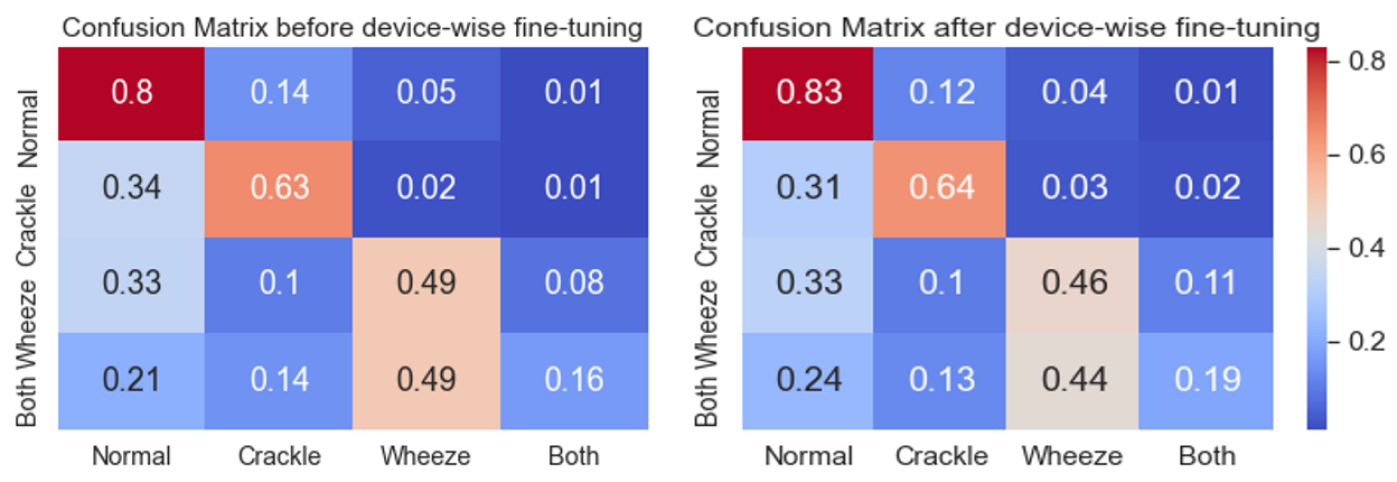
Device specific fine-tuning: We found that the large skew in sample distribution across devices caused the model to not generalize well for under-represented devices. Our device specific fine-tuning scheme helped significantly, resulting in an improvement of in the final ICBHI score. We also observed that this fine-tuning disproportionally helped the under-represented classes. Table 3 shows that devices with fewer samples had 9% increase in their scores.
| Device | % Samples | Score Improvement |
|---|---|---|
| AKGC417L | 63% | 1.7% |
| Meditron | 21% | 1.6% |
| Litt3200 | 9% | 9.3% |
| LittC2SE | 7% | 8.6% |
4 Related Work
Recently, there has been a lot of interest in using deep learning models for respiratory sounds classification [1, 12, 9]. It has outperformed statistical methods (HMM-GMM) [8] and traditional machine learning methods (boosted decision trees, SVM) [4, 24]. In these deep learning based approaches, a time-frequency representation of the audio signal is provided as input to the model. Kochetov et al. [9] propose a deep recurrent network with a noise masking intermediate step for the four class classification task, obtaining a score of on the 80-20 split. However the paper omits the details regarding noise label generation [1], thus making it hard to reproduce. Deep residual networks and optimized S-transform based features are used by Chen et al. [6] for three-class classification of anomalies in lung sounds. The model is trained and tested on a smaller subset of the ICBHI dataset on a 70-30 split and achieve a score of .
Acharya and Basu [1] propose a Mel-spectrogram based hybrid CNN-RNN model with patient-specific model tuning, achieving a score of on 4-class and 80-20 split. Ma et al. [12] introduce LungRN+NL which incorporates a non-local block in the ResNet architecture and apply mixup augmentations to address the data imbalance problem and improve the model’s robustness, achieving sensitivity of . However, none of these approaches focus on characteristics of the ICBHI dataset, which we exploit to improve performance.
5 Conclusion and Future Work
The paper proposes RespireNet a simple CNN-based model, along with a set of novel techniques—device specific fine-tuning, concatenation-based augmentation, blank region clipping, and smart padding—enabling us to effectively utilize a small-sized dataset for accurate abnormality detection in lung sounds. Our proposed method achieved a new SOTA for the ICBHI dataset, on both the 2-class and 4-class classification tasks. Further, our proposed techniques are orthogonal to the choice of network architecture and should be easy to incorporate within other frameworks.
The current performance limit of the 4-class classification task can be mainly attributed to the small size of the ICBHI dataset, and the variation among the recording devices. Furthermore, there is lack of standardization in the 80-20 split and we found variance in the results based on the particular split. In future, we would recommend that the community should focus on capturing a larger dataset, while taking care of the issues raised in this paper.
References
- Acharya and Basu [2020] Jyotibdha Acharya and Arindam Basu. Deep neural network for respiratory sound classification in wearable devices enabled by patient specific model tuning. IEEE Transactions on Biomedical Circuits and Systems, PP:1–1, 03 2020.
- Aykanat et al. [2017] Murat Aykanat, Özkan Kiliç, Bahar Kurt, and S. Saryal. Classification of lung sounds using convolutional neural networks. EURASIP Journal on Image and Video Processing, 2017:1–9, 2017.
- Bohadana et al. [2014] Abraham Bohadana, Gabriel Izbicki, and Steve Kraman. Fundamentals of lung auscultation. The New England journal of medicine, 370:744–751, 02 2014.
- Chambres et al. [2018] Gaetan Chambres, Pierre Hanna, and Myriam Desainte-Catherine. Automatic detection of patient with respiratory diseases using lung sound analysis. pages 1–6, 09 2018.
- Chattopadhyay et al. [2018] A. Chattopadhyay, Anirban Sarkar, Prantik Howlader, and V. Balasubramanian. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), pages 839–847, 2018.
- Chen et al. [2019] Hai Chen, Zhiyuan Pei, Mianjie Li, and Jianqing Li. Triple-classification of respiratory sounds using optimized s-transform and deep residual networks. IEEE Access, PP:1–1, 03 2019.
- Flietstra et al. [2011] B Flietstra, Natasha Markuzon, Andrey Vyshedskiy, and R Murphy. Automated analysis of crackles in patients with interstitial pulmonary fibrosis. Pulmonary medicine, 2011:590506, 01 2011.
- Jakovljevic and Loncar-Turukalo [2018] Niksa Jakovljevic and Tatjana Loncar-Turukalo. Hidden Markov Model Based Respiratory Sound Classification, pages 39–43. 01 2018.
- Kochetov et al. [2018] Kirill Kochetov, Evgeny Putin, Maksim Balashov, Andrey Filchenkov, and Anatoly Shalyto. Noise Masking Recurrent Neural Network for Respiratory Sound Classification: 27th International Conference on Artificial Neural Networks, Rhodes, Greece, October 4–7, 2018, Proceedings, Part III, pages 208–217. 10 2018. ISBN 978-3-030-01423-0.
- Liu et al. [2019] Renyu Liu, Shengsheng Cai, Kexin Zhang, and Nan Hu. Detection of adventitious respiratory sounds based on convolutional neural network. pages 298–303, 11 2019.
- Ma et al. [2019] Yi Ma, Xinzi Xu, Qing Yu, Yuhang Zhang, Yongfu Li, Jian Zhao, and Guoxing Wang. Lungbrn: A smart digital stethoscope for detecting respiratory disease using bi-resnet deep learning algorithm. 10 2019.
- Ma et al. [2020] Yi Ma, Xinzi Xu, and Yongfu Li. Lungrn+nl: An improved adventitious lung sound classification using non-local block resnet neural network with mixup data augmentation. 08 2020.
- Messner et al. [2018] Elmar Messner, Melanie Fediuk, Paul Swatek, Stefan Scheidl, Freyja-Maria Smolle-Juttner, Horst Olschewski, and Franz Pernkopf. Crackle and breathing phase detection in lung sounds with deep bidirectional gated recurrent neural networks. volume 2018, pages 356–359, 07 2018.
- Organization [2017] World Health Organization. The global impact of respiratory diseases (2nd edition). Forum of International Respiratory Societies (FIRS), 2017.
- Perna and Tagarelli [2019] D. Perna and A. Tagarelli. Deep auscultation: Predicting respiratory anomalies and diseases via recurrent neural networks. 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), pages 50–55, 2019.
- Perna [2018] Diego Perna. Convolutional neural networks learning from respiratory data. pages 2109–2113, 12 2018.
- Polat and Guler [2005] Hüseyin Polat and Inan Guler. A simple computer-based measurement and analysis system of pulmonary auscultation sounds. Journal of medical systems, 28:665–72, 01 2005.
- Pramono et al. [2017] Renard Xaviero Adhi Pramono, Stuart A. Bowyer, and E. Rodríguez-Villegas. Automatic adventitious respiratory sound analysis: A systematic review. PLoS ONE, 12, 2017.
- Reichert et al. [2008a] Sandra Reichert, Gass Raymond, Christian Brandt, and Emmanuel Andrès. Analysis of respiratory sounds: State of the art. Clinical Medicine : Circulatory, Respiratory and Pulmonary Medicine, 2, 05 2008a.
- Reichert et al. [2008b] Sandra Reichert, Gass Raymond, Christian Brandt, and Emmanuel Andrès. Analysis of respiratory sounds: State of the art. Clinical Medicine : Circulatory, Respiratory and Pulmonary Medicine, 2, 05 2008b.
- Rocha et al. [2018] B. M. Rocha, D. Filos, L. Mendes, I. Vogiatzis, E. Perantoni, E. Kaimakamis, P. Natsiavas, A. Oliveira, C. Jácome, A. Marques, R. P. Paiva, I. Chouvarda, P. Carvalho, and N. Maglaveras. respiratory sound database for the development of automated classification. In Nicos Maglaveras, Ioanna Chouvarda, and Paulo de Carvalho, editors, Precision Medicine Powered by pHealth and Connected Health, pages 33–37, Singapore, 2018. Springer Singapore.
- Rocha et al. [2019] Bruno Rocha, D. Filos, Luís Mendes, Gorkem Serbes, Sezer Ulukaya, Yasemin Kahya, Niksa Jakovljevic, Tatjana Loncar-Turukalo, Ioannis Vogiatzis, Eleni Perantoni, Evangelos Kaimakamis, Pantelis Natsiavas, Ana Oliveira, Cristina Jácome, Alda Marques, N. Maglaveras, Rui Pedro Paiva, Ioanna Chouvarda, and Paulo De Carvalho. An open access database for the evaluation of respiratory sound classification algorithms. Physiological Measurement, 40, 02 2019.
- Russakovsky et al. [2015] Olga Russakovsky, Jia Deng, Hao Su, Jonathan Krause, Sanjeev Satheesh, Sean Ma, Zhiheng Huang, Andrej Karpathy, Aditya Khosla, Michael Bernstein, Alexander C. Berg, and Li Fei-Fei. ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision (IJCV), 115(3):211–252, 2015.
- Serbes et al. [2018] Gorkem Serbes, Sezer Ulukaya, and Yasemin Kahya. An Automated Lung Sound Preprocessing and Classification System Based OnSpectral Analysis Methods, pages 45–49. 01 2018.
- Shi et al. [2019] Lukui Shi, Kang Du, Chaozong Zhang, Hongqi Ma, and Wenjie Yan. Lung sound recognition algorithm based on vggish-bigru. IEEE Access, PP:1–1, 09 2019.
6 Supplementary Material
This supplementary material includes some other details about the dataset, and additional results which could not be accommodated in the main paper.
6.1 Dataset Details
The 2017 ICBHI dataset [21] comprises of 920 recordings from 126 patients with a combined total duration of 5.5 hours. Each breathing cycle in a recording is annotated by a single expert as one of the four classes: normal, crackle, wheeze or both (crackle and wheeze). These cycles have various recording lengths (see Figure 4) ranging from 0.2s to 16.2s (mean cycle length is 2.7s) and the number of cycles is imbalanced across the four classes (i.e. 3642, 1864, 886, 506 cycles for normal, crackle, wheeze and both classes respectively).

The dataset consists of sound recordings from four devices AKGC417L Microphone, 3M Littmann Classic II SE Stethoscope, 3M Litmmann 3200 Electronic Stethoscope and WelchAllyn Meditron Master Elite Electronic Stethoscope and is not balanced across patients as well as number of breathing cycles (see Tables 4, 5). This creates a skew in the data distribution and has an adverse impact on the performance of the model as discussed in the analysis earlier.
| Device | Patient Count* | N | C | W | B | Total |
|---|---|---|---|---|---|---|
| AKGC417L | 32 | 1922 | 1543 | 500 | 381 | 4346 |
| Meditron | 64 | 1037 | 215 | 148 | 56 | 1456 |
| Litt3200 | 11 | 347 | 77 | 126 | 44 | 594 |
| LittC2SE | 23 | 336 | 29 | 112 | 25 | 502 |
*Number of patients total to 130 instead of 126 as some of the devices have an overalap with the patients.
| Device | N | C | W | B |
|---|---|---|---|---|
| AKGC417L | 0.53 | 0.83 | 0.56 | 0.75 |
| Meditron | 0.28 | 0.11 | 0.17 | 0.11 |
| Litt3200 | 0.10 | 0.02 | 0.14 | 0.09 |
| LittC2SE | 0.09 | 0.04 | 0.13 | 0.05 |
For creating the splits we perform sample 80-20 w.r.t number of patients. From the numbers in Table 4, we have 64 patients from Meditron device but only 1468 breathing cycles (22.9 breathing cycles per patient on an average), whereas for AKGC417L device we have 32 patients and 4364 breathing cycles (136.4 breathing cycles per patient on an average). This depicts the huge skew in the splits across devices and patients. Further there is also a skew between abnormal classes across devices: The majority of crackle class (83% of the total samples) is found within the AKGC417L device whereas wheeze and both have different proportions across devices.
6.2 Additional Results
Single Device Training We train our model only on samples from the AKGC417L device. Table 6 depicts the test performance on the 4 different devices. This demonstrates that the training only on a single device, does not translate well across the other devices, thus further motivating the use of device specific fine-tuning.
| Device | Normal | Crackle | Wheeze | Both |
|---|---|---|---|---|
| AKGC417L | 61.3% | 77.5% | 23.8% | 28.2% |
| Meditron | 47.3% | 69.2% | 26.3% | 0.0% |
| Litt3200 | 51.2% | 66.7% | 20.7% | 66.7% |
| LittC2SE | 16.8% | 22.2% | 0.0% | 0.0% |
Attention Map Visualization Figure 5 depicts global average of attention maps computed (for layer-4 of ResNet34) using Grad-Cam++[5] for 1370 samples in the test split before and after employing the blank region clipping scheme during network training. It can be observed that the network starts focusing more on the bottom half of the spectrogram, instead of blank spaces after using blank region clipping. This demonstrates the efficacy of using the proposed blank region clipping scheme which also results in improved performance.

Confusion Matrix Figure 6 shows the confusion matrix before and after device specific fine-tuning.