QCResUNet: Joint Subject-level and Voxel-level Segmentation Quality Prediction
Abstract
Deep learning has made significant strides in automated brain tumor segmentation from magnetic resonance imaging (MRI) scans in recent years. However, the reliability of these tools is hampered by the presence of poor-quality segmentation outliers, particularly in out-of-distribution samples, making their implementation in clinical practice difficult. Therefore, there is a need for quality control (QC) to screen the quality of the segmentation results. Although numerous automatic QC methods have been developed for segmentation quality screening, most were designed for cardiac MRI segmentation, which involves a single modality and a single tissue type. Furthermore, most prior works only provided subject-level predictions of segmentation quality and did not identify erroneous parts segmentation that may require refinement. To address these limitations, we proposed a novel multi-task deep learning architecture, termed QCResUNet, which produces subject-level segmentation-quality measures as well as voxel-level segmentation error maps for each available tissue class. To validate the effectiveness of the proposed method, we conducted experiments on assessing its performance on evaluating the quality of two distinct segmentation tasks. First, we aimed to assess the quality of brain tumor segmentation results. For this task, we performed experiments on one internal (Brain Tumor Segmentation (BraTS) Challenge 2021, ) and two external datasets (BraTS Challenge 2023 in Sub-Saharan Africa Patient Population (BraTS-SSA), ; Washington University School of Medicine (WUSM), ). Specifically, we first performed a three-fold cross-validation on the internal dataset using segmentations generated by different methods at various quality levels, followed by an evaluation on the external datasets. Second, we aimed to evaluate the segmentation quality of cardiac Magnetic Resonance Imaging (MRI) data from the Automated Cardiac Diagnosis Challenge (ACDC, ). The proposed method achieved high performance in predicting subject-level segmentation-quality metrics and accurately identifying segmentation errors on a voxel basis. This has the potential to be used to guide human-in-the-loop feedback to improve segmentations in clinical settings.
1 Introduction
Medical image segmentation plays an indispensable role for accurate diagnosis, monitoring, treatment planning, and population studies of diseases in modern medicine by enabling precise delineation of anatomical structures and pathological regions (Garcia-Garcia et al., 2017; Litjens et al., 2017). In particular, precise segmentation of healthy and abnormal anatomy into multiple classes using magnetic resonance imaging (MRI) is crucial in this process. Recently, deep learning-based methods have achieved state-of-the-art performance in automated segmentation tasks including brain tumor (Ronneberger et al., 2015; Kamnitsas et al., 2017; Isensee et al., 2018a, b; Baid et al., 2021) and cardiac (Tran, 2016; Khened et al., 2019; Zhou et al., 2021) MRI segmentation. However, deep neural networks are sensitive to data distribution. This makes them prone to reduced performance when applied on out-of-distribution MRI scans due to variations in acquisition protocols, contrast, image quality, etc. Therefore, quality control (QC) is necessary to thoroughly assess segmentations before they are used for clinical purposes or large-scale research studies. QC tools are required to detect severe segmentation failures on a per-case basis, pinpoint areas needing segmentation refinement at the voxel level, and provide a quality measure for downstream analyses. Previously proposed QC methods fall into four main categories: uncertainty estimation-based, generative model-based, reverse classification accuracy (RCA)-based, and regression-based.
The first category of automated segmentation QC methods operates on the premise that high uncertainty is reflective of poor quality segmentations (Ng et al., 2018; Albà et al., 2018; Sander et al., 2020; Ng et al., 2020; Bai et al., 2018; Roy et al., 2018; Jungo et al., 2020; Mehta et al., 2022). Accordingly, most studies focused on developing uncertainty measures to aggregate voxel-wise uncertainty as a proxy for segmentation quality measures, such as Dice Similarity Coefficient (DSC). However, most developed proxy measures (Ng et al., 2018; Roy et al., 2018; Jungo et al., 2020) have not demonstrated a strong correlation with DSC. Moreover, non-negligible errors in uncertainty estimation were observed at the voxel level resulting in unreliable subject-level uncertainty aggregation (Jungo et al., 2020). Additionally, these methods can only be applied to deep learning-based models, and thus cannot be used to assess the quality of segmentations obtained by other methods.
The second category of automated QC methods is based on the assumption that there is a relationship between image intensities and tissue labels. Along that direction, Grady et al. (2012) proposed a Gaussian mixture model with hand-crafted features (i.e., geometric features, intensity features, gradient features, and ratio features) to characterize the segmentation quality and detect segmentation failure. Wang et al. (2020b) proposed a variational autoencoder (VAE) to learn the latent representation of pairs of images and their ground-truth segmentations. During inference, the encoder is frozen while the decoder is refined for a given image-segmentation pair to produce a surrogate segmentation. The subject-level DSC is then computed between the query segmentation and the surrogate segmentation. The tuning of the decoder is required for each query image, which may be computationally expensive and time-consuming. Leveraging the advances in the image-to-image translation task, Li et al. (2022) proposed a generative adversarial network (GAN) to generate an informative reference image conditioned on the query segmentation mask that needs to be assessed. An auxiliary network (i.e., difference investigator) is then trained to predict image-level and pixel-level quality by taking the raw input image and the generated reference image as inputs. These approaches have only been validated in cardiac MRI segmentation QC that involves only a single modality and segmentation with regular shapes. Translating it to the more complex brain tumor segmentation QC scenario is difficult due to the presence of multiple modalities and intratumoral tissue heterogeneities, which are challenging to model.
The third category of the automated segmentation QC methods is built upon the reverse classification accuracy (RCA) framework (Valindria et al., 2017). This was initially designed for whole-body multi-organ MRI segmentation QC and was later applied to cardiac MRI datasets (Robinson et al., 2019). The RCA framework involves: (i) choosing a reference dataset with known ground-truth segmentation, (ii) training a segmenter using a query image-segmentation pair, (iii) using the trained segmenter to segment the images in the reference dataset, and (iv) estimating DSC as the maximum DSC achieved by the trained segmenter in the reference dataset. Although effective for whole-body multi-organ MRI segmentation and cardiac MRI segmentation QC, RCA’s reliance on a representative reference dataset and image registration poses challenges in brain tumors. The large variability in brain tumors makes the reference dataset hard to be representative. Different from whole-body multi-organ and cardiac segmentation, which have consistent shapes and appearances, the heterogeneous appearance and phenotypes of brain tumors complicate the establishment of correspondences for the tumorous areas between different subjects. Lastly, despite predicting subject-level DSC, the RCA framework lacks segmentation error localization at the voxel level.
The fourth category of the segmentation QC methods is regression-based methods that directly predict the subject-level DSC. Early attempts employed a Support Vector Machine regression in combination with hand-crafted features to detect cardiac MRI segmentation failures (Kohlberger et al., 2012; Albà et al., 2018). Instead of using hand-crafted features, Robinson et al. (2018) proposed a convolutional neural network (CNN) regressor to automatically extract features from a large cardiac MRI segmentation dataset to predict DSC. Besides predicting DSC, Kofler et al. (2022) proposed a holistic rating to approximate how expert neuroradiologists classify high-quality and poor-quality segmentations. However, manually deriving holistic ratings by clinical experts is laborious and prone to inter-rater variability, hindering its applicability in large-scale datasets. Despite the fact that satisfactory performance was achieved in predicting subject-level segmentation metrics by these regression-based methods, voxel-level localization of segmentation error is unavailable.
Although numerous efforts have been devoted to automated segmentation QC, the majority of the studies have focused on whole-body multi-organ MRI segmentation and cardiac MRI segmentation QC. Additionally, most of these approaches have not assessed out-of-distribution generalization. Importantly, there has been limited research on segmentation QC specifically for brain tumor MRI segmentation QC. The heterogeneous and complex appearances of brain tumors, which vary in locations, sizes, and shapes, contribute to the increased challenge of QC in brain tumor MRI segmentation. In addition, most prior QC approaches focused on predicting subject-level DSC (Jungo et al., 2020; Robinson et al., 2018, 2019; Valindria et al., 2017; Li et al., 2022) and do not consider the quality of the segmentation contour. However, DSC and contour measurement (e.g., normalized surface dice (NSD)) are equally important to comprehensively assess the segmentation quality (Maier-Hein et al., 2024). Furthermore, it is crucial to address reliable voxel-level tissue-specific segmentation error localization. This localization is vital not only for auditing purposes but also for radiologists to prioritize cases that require manual refinement. By incorporating this component into quality control measures, it can enhance the accuracy and effectiveness of the overall segmentation process. However, this aspect has been largely overlooked in previous studies. One exception is the work proposed by Li et al. (2022). However, this approach was only validated on cardiac segmentation QC with limited datasets. In addition, this method can only provide a binary segmentation error mask and is unable to identify voxel-level segmentation errors for different tissue classes.

To address these limitations, we proposed a novel deep learning model, termed QCResUNet, to jointly predict segmentation-quality metrics at the subject level and localize segmentation errors across different tissue classes at the voxel level. This work extends our previous preliminary work (Qiu et al., 2023) in several ways. First, we extended the previous work to predict subject-level DSC and NSD as well as a collection of binary segmentation error maps, each corresponding to a different tissue class, to provide a more comprehensive evaluation of segmentation quality. To achieve this goal, we further proposed an attention-based segmentation error map aggregation mechanism to better delineate segmentation errors for different tissue classes. Second, we conducted rigorous evaluations to validate the generalizability of the proposed QCResUNet by performing a three-fold cross-validation on the internal dataset and evaluating the proposed method on out-of-distribution datasets with MRI scans of varying image quality and segmentations produced. Third, we further examined the generalizability of the proposed QCResUNet by evaluating its ability to assess the segmentation quality for cardiac MRI. Fourth, we extensively evaluated the performance of the proposed method in comparison with several state-of-the-art segmentation QC methods, including RCA-based (Valindria et al., 2017) and uncertainty-based methods (Jungo et al., 2020). Fifth, we performed an in-depth explainability analysis to elucidate the reasons behind the proposed method’s superior performance compared to other competing approaches.
The main contributions of this work are fourfold:
-
1.
We proposed a multi-task learning framework named QCResUNet to simultaneously predict the DSC and NSD at the subject level and localize segmentation errors at the voxel level.
-
2.
We proposed an attention-based mechanism to aggregate the segmentation error map that better handles voxel-level segmentation error prediction for different tissue classes.
-
3.
We extensively evaluated the performance of the proposed model using both internal and external testing for the brain tumor segmentation QC task. Internal training, validation, and testing were performed through a three-fold cross-validation on 1251 cases from Brain Tumor Segmentation (BraTS 2021). External testing was performed on independent datasets from Washington University School of Medicine (WUSM) and BraTS Challenge 2023 in Sub-Saharan Africa Patient Population (BraTS-SSA). Our results demonstrated that the proposed model can generalize well on the out-of-distribution cases from different brain tumor datasets that have been segmented by various methods.
-
4.
The proposed model was also evaluated on the cardiac MRI segmentation QC task, demonstrating its potential for application in a broader range of QC tasks beyond brain tumor segmentation QC.
2 Materials and methods
2.1 Dataset
In this study, we used three datasets for evaluation of the QC performance on brain tumor segmentation. First, we used pre-operative multimodal MRI scans with gliomas of all grades (WHO Central Nervous System grades 2-4) grades from the BraTS 2021 challenge training dataset (). The BraTS dataset is a heterogeneous dataset consisting of cases from 23 different sites with various levels of quality and protocols. The BraTS dataset was used for training, validation, and internal testing. Additionally, we used two datasets that are not included in the BraTS 2021 dataset (i.e., the BraTS-SSA dataset and the WUSM dataset) for external testing, allowing for an unbiased assessment of the generalizability of the proposed method. The BraTS-SSA dataset () (Adewole et al., 2023) is an extension of the original BraTS 2021 dataset with patients from Sub-Saharan Africa, which includes lower-quality MRI scans (e.g., poor image contrast and resolution) as well as unique characteristics of gliomas (i.e., suspected higher rates of gliomatosis cerebri). The WUSM dataset () was obtained from the retrospective health records of the Washington University School of Medicine (WUSM), with a waiver of consent, in accordance with the Health Insurance Portability and Accountability Act, as approved by the Institutional Review Board (IRB) of WUSM (IRB no. PA18-1113). Each subject in all datasets comprised four modalities viz. pre-contrast T1-weighted (T1), T2-weighted (T2), post-contrast T1-contrast (T1c), and Fluid attenuated inversion recovery (FLAIR). In addition, multi-class tumor segmentation masks annotated by experts were also available. Segmentation masks delineated enhancing tumor (ET), non-enhancing tumor core (NCR), and edema (ED) classes. Following standard BraTS procedures, we combined the binary ET, NCR and ED segmentation masks to delineate the whole tumor (WT), tumor core (TC), and enhancing tumor. The WT mask consists of all tumor tissue classes (i.e., ET, NCR, and ED), while the TC mask comprises ET and NCR tissue classes.
Scans from the BraTS training and BraTS-SSA datasets were already registered to the SRI24 anatomical atlas (Rohlfing et al., 2010), resampled to 1-mm3 isotropic resolution and skull-stripped. For consistency, raw MRI scans from WUSM were pre-processed following the same protocol using the Integrative Imaging Informatics for Cancer Research: Workflow Automation for Neuro-oncology (Chakrabarty et al., 2022) framework. Subsequently, we z-scored all the skull-stripped scans in the BraTS datasets and the WUSM dataset on a per-scan basis. Finally, scans from the entire dataset were cropped to exclude background regions and then were zero-padded to a common dimension of using the nnUNet preprocessing pipeline (Isensee et al., 2018b).
In evaluating the QC performance on the cardiac segmentation task, we used the Automated Cardiac Diagnosis Challenge (ACDC) dataset (Bernard et al., 2018), which consists of 100 subjects (200 volumes). Each volume in the ACDC dataset is associated with a multi-class segmentation mask delineating the left ventricle (LV), myocardium (Myo), and right ventricle (RV). Lastly, each volume was cropped and zero-padded to a common dimension of using the nnUNet pipeline (Isensee et al., 2018b).
2.2 Segmentation Dataset Generation
The majority of previous regression-based methods were trained and validated on datasets limited by both sample size and heterogeneity. To address this limitation, we constructed an extensive dataset consisting of segmentation results of diverse quality levels. This enabled us to capture the variability of segmentations and obtain a reliable performance assessment of the proposed method on a wide range of segmentations. For this purpose, we adopted a four-step approach to produce segmentation results using different methods with various combinations of imaging modalities as input.
As far as the brain tumor segmentation QC task is concerned, we first employed both CNN-based (i.e., nnUNet (Isensee et al., 2018b)) and Transformer-based (i.e., nnFormer (Zhou et al., 2021)) segmentation UNets and trained them independently seven times by taking different modalities as input (namely T1-only, T1c-only, T2-only, FLAIR-only, T1-FLAIR, T1c-T2, and all four modalities). Since each modality is sensitive to certain tissue types (e.g., T1c is effective in detecting enhancing tumors and FLAIR is advantageous in identifying edema), this approach enabled us to produce segmentations with varying levels of quality. Second, we sampled the segmentation along the training routines at various iterations to generate segmentation results from both fully trained and inadequately trained models. For this purpose, a small learning rate () was used during training to slower their convergence, allowing the segmentation to improve gradually from low quality to high quality. Third, we employed a data augmentation strategy based on image transformations to produce diverse segmentation results, termed SegGen. A series of image transformations, including rotation, translation, scaling, and elastic deformation, were randomly applied to each ground-truth segmentation three times with a probability of (see Table 1). Fourth, apart from the segmentations produced by the aforementioned three methods, we also created out-of-sample segmentation samples from a different segmentation method to assess the generalizability of our proposed model in the testing phase. To accomplish this, we employed the DeepMedic framework (Kamnitsas et al., 2017) to train seven segmentation models using the same diverse input modalities as we did in training the nnUNet and nnFormer. After applying the aforementioned four steps, we were able to create a set of erroneous segmentations for the cases included in the internal and external datasets. The erroneous segmentations along with the ground truth segmentations were subsequently used to estimate the DSC, NSD, and the ground truth segmentation error map (SEM) that were used to train the proposed network. The SEM was computed as the difference between a query segmentation (i.e., one of the erroneous segmentations we produced) and the corresponding ground truth. Examples of the generated segmentations can be found in Figure 1(b).
For the cardiac segmentation task, we employed a procedure similar to the one used in generating segmentation for the brain tumor task. However, unlike brain tumor MRI, cardiac MRI involves only a single modality. Consequently, we trained one nnUNet model and one nnFormer model, sampling the segmentation along the training routines at various iterations. We also generated segmentation results using SegGen during model training. However, due to the technical difficulty of training DeepMedic on the ACDC dataset (i.e., one axis in the ACDC dataset has a significantly smaller input size), we could not include any segmentation produced by DeepMedic for the cardiac segmentation QC task.
| Transformation | Parameters | Probability |
| Rotation | [, ] | 0.5 |
| Scaling | scales=[, ] | 0.5 |
| Translation | moves=[, ] | 0.5 |
| Deformation | displacements=[-20, 20] | 0.5 |

Despite our efforts to sample across different convergence stages by adopting a small learning rate, we observed that the deep learning models could successfully segment the majority of the cases. As a consequence, the resulting dataset was distributed unevenly across the different levels of quality, comprising mostly of higher quality segmentation results (see Figure 1(a)). Following the resampling strategy in Robinson et al. (2018), we randomly selected a subset of samples from each bin of the DSC histogram. This ensured that the number of segmentations from each bin was equal to the lowest count-per-bin value () across the distribution for all the segmentation datasets (refer to Figure 1(a)). For this purpose, we divided DSC values into 10 evenly spanned bins ranging from 0 to 1. We sampled cases from these bins such that we ensured that all bins contained an equal number of cases. For the training set, we did not manually select the DSC samples. Instead, at each training iteration, we randomly sampled segmentations that were evenly distributed across 10 bins to avoid imbalanced DSC values in each training batch. We kept this process stochastic to let the model see as many samples as possible without skewing its exposure to either low DSC or high DSC samples. This means we randomly sampled segmentations for bins that contained more than samples at each training iteration. Conversely, during the evaluation phase, we opted for a deterministic resampling approach, which enabled unbiased evaluations across various quality levels. In order to account for the variability introduced by the resampling process, we implemented a three-fold cross-validation strategy for all our experiments (see Section 3).
2.3 QCResUNet
The proposed 3D U-shaped QCResUNet (Figure 2(a)) aims to automatically evaluate the quality of a query multi-class segmentation mask by predicting subject-level segmentation quality measures as well as identifying voxel-level segmentation error maps for each tissue class (SEMs). Without loss of generality, QCResUNet takes as input imaging modalities represented by (e.g., the brain tumor segmentation QC case) and the query multi-class segmentation mask (). The outputs of QCResUNet include two complementary subject-level segmentation quality measures (DSC and NSD) and a collection of binary voxel-level SEMs, each delineating the segmentation error corresponding to a specific tissue class in . We would like to point out that we followed the standard definition of DSC for calculating the subject-level DSC for multi-class segmentation masks (see A).
2.3.1 Network Design
The U-shaped QCResUNet is designed to perform both regression and segmentation tasks. Therefore, the proposed QCResUNet consists of three parts that are trained end-to-end: (i) a ResNet-34 encoder for DSC and NSD prediction; (ii) a decoder architecture for predicting the multiclass SEM, and (iii) an attention-based SEM aggregation module.
Encoder: For the purpose of predicting subject-level DSC, we adopted a ResNet-34 (He et al., 2016) architecture as part of the encoding path of our network, which can capture semantically rich features that are important for accurately characterizing segmentation quality. While retaining the main structure of the 2D ResNet-34 in He et al. (2016), we made the following modifications to account for the 3D nature of the input data. First, all 2D convolutional and pooling layers were replaced by 3D counterparts (see Figure 2(b)). Second, we replaced all batch normalization blocks (Ioffe and Szegedy, 2015) with instance normalization blocks (Ulyanov et al., 2016) to cater to the small batch size during 3D model training. Third, to prevent overfitting, spatial dropout (Tompson et al., 2015) was added to each residual block with a probability of 0.3 to randomly zero out channels in the feature map (see Figure 2(b)).
Decoder: To estimate a segmentation error map with high spatial resolution and accurate localization information, we designed a decoder architecture. The decoder takes the low-resolution contextual features of segmentation quality that were extracted by the encoder and transfers them to a high-resolution multiclass SEM. This was accomplished by first upsampling the input feature map by a factor of two using nearest neighbor interpolation, followed by a convolutional layer. Next, we concatenated the upsampled feature maps with the corresponding encoder level’s features using skip-connections to facilitate information flow from the encoder to the decoder. This was followed by two convolutional blocks that reduced the number of feature maps back to the original value before concatenation. Each convolutional block comprised a convolutional layer, followed by an instance normalization layer and a Leaky Rectified Linear Unit (LeakyReLU) activation function (Maas et al., 2013) (see Figure 2(c)).
Importantly, we used the middle-level semantics in the intermediate block (Block 3 in Figure 2(a)) of the ResNet encoder as the input to the decoder. The rationale behind this was that the last block (Block 4 in Figure 2(a)) of the ResNet encoder contains features that are specific to the DSC prediction task. In contrast, the middle-level features are likely to constitute a more universal semantic representation that characterizes the segmentation error (Ahn et al., 2019).
Attention-based SEM aggregation: To better predict the segmentation error map corresponding to different tissue classes (e.g., [, , ] in the brain tumor segmentation QC task), we propose an attention-based SEM aggregation mechanism. This is because each tissue class in the query segmentation mask () contributes differently to the predicted SEM. Specifically, we leveraged the efficient channel attention (ECA) (Wang et al., 2020a) to model the correlation between the features in the last layer of the decoder and the one-hot encoded query segmentation mask. A convolution layer was then applied to the output of the ECA module to predict tissue-level SEMs (Figure 2(a)). Specifically, the channel attention was computed by feeding the pooled input feature (after the global average pooling layer) to a 1D convolutional layer followed by a Sigmoid activation (Figure 2(d)).
2.3.2 Network configuration
The overall model configuration used for both the brain tumor and cardiac segmentation QC tasks was identical to the one outlined in Figure 2. The only exception is that we used different downsampling/upsampling rates for the brain tumor and cardiac cases to accommodate differences in their respective input sizes. Specifically, the dowsampling/upsampling rates used for the brain tumor case were [[2, 2, 2], [2, 2, 2], [2, 2, 2], [2, 2, 2]] with each [2, 2, 2] represents the downsampling/upsampling rates for axial, sagittal, and coronal axes at each stage. In contrast, the dowsampling/upsampling rates used for the cardiac case were [[1, 2, 2], [1, 2, 2], [1, 2, 2], [2, 2, 2]]. This is because the first axis in the ACDC dataset is ten times smaller than the other two axes.
2.3.3 Multi-task learning objective
The proposed QCResUNet was jointly trained for predicting DSC, NSD, and SEM by optimizing the combination of a mean absolute error (MAE) loss, a DSC loss, and a cross-entropy (CE) loss.
The DSC and NSD prediction task was trained by minimizing the MAE loss ():
where is the total number of samples in the training dataset, and is indexing each sample. The MAE loss () quantifies the dissimilarities between the ground truth DSC/NSD (DSCgt/NSDgt) and the predicted DSC/NSD (DSCpred/NSDpred). Compared to the mean squared error loss commonly used for regression tasks, the MAE loss has been proved to be more sensitive to outliers (Qi et al., 2020). This enhances the robustness of QCResUNet to outliers.
The voxel-level segmentation error prediction task was trained by optimizing both the DSC and CE losses. Though there are many variants of the dice loss, we opted for the one proposed in (Drozdzal et al., 2016) due to its wide success in a variety of medical imaging segmentation tasks:
where is the total number of voxels in a batch, and indexes each voxel; is the total number of tissue classes, and indexes each class. Here, SEM and SEM refer to the ground truth for the tissue class SEM and its probabilistic prediction generated from the decoder’s sigmoid output, respectively. The CE loss is defined as
The average of the dice loss and CE loss was performed over the total number of voxels () present in a batch.
To balance the loss for the subject-level DSC and NSD prediction task and the voxel-level SEM prediction task, we combined the three losses in a weighted fashion. The final objective function is given as
where is the loss balance parameter.
3 Experimental Validation
We evaluated the proposed method on both brain tumor and cardiac MRI segmentation QC tasks to demonstrate its effectiveness and generalizability across different datasets and tasks.
3.1 Brain tumor MRI segmentation QC task
We performed three-fold cross-validation to validate the effectiveness of the proposed method for the brain tumor segmentation QC task. For this purpose, we held out an internal testing set with 251 subjects. In each iteration of the three-fold cross-validation, the remaining BraTS dataset was randomly partitioned into training and validation subsets with 667 and 333 subjects, respectively. The application of the previously described four-step approach and resampling (see Section 2.2) resulted in 98,048 training segmentation samples, 28,800 validation segmentation samples, as well as 39,063 internal testing segmentations (36,144 from nnUNet and nnFormer, 2,919 from DeepMedic). We only used the segmentations generated by the DeepMedic as the testing set. This is because segmentations generated by DeepMedic were not seen during training and validation. The BraTS-SSA and WUSM datasets were used as external testing datasets to validate the generalizability of the proposed method. By employing the previously described four-step approach (see Section 2.2), we extended the original BraTS-SSA, and WUSM datasets to include 6,040 and 26,425 segmentations, respectively. After applying the resampling in Section 2.2, the internal BraTS testing, BraTS-SSA, and WUSM datasets were resampled to include 4,753, 1,204, and 2,693 segmentations, respectively.
We performed hyperparameter tuning to determine two critical hyperparameters in the proposed method: the initial learning rate and the loss balance parameter . Hyperparameter tuning was carried out by performing a Bandit-based Bayesian optimization on the training and validation dataset implemented using the Raytune framework (Liaw et al., 2018). Specifically, we sampled the learning rate from a range of following a log-uniform distribution. Additionally, the loss balance parameter was chosen from the following set of values . The optimal balance parameter for QCResUNet was determined to be 1. However, we did not observe a significant difference when varying from 0.1 to 1 and 2 (see Figure S1). Although the optimal learning rate varied from model to model (refer to Table 2), it generally fell within the scale of as we used a relatively small batch size of 4. We kindly direct the readers to C for the detailed learning curve in hyperparameter tuning using RayTune.
Results reported hereafter were obtained using the above optimal hyper-parameters. For both internal (BraTS) and external (BraTS-SSA and WUSM) testing sets, we reported the results after performing a model ensemble technique for the proposed method and all baseline methods. This involved averaging the results from three models for DSC and NSD prediction and performing majority voting for the SEM prediction. The mean ( standard deviation) of MAEs and DSC were calculated over all subjects.
| Method | learning rate | balance parameter () |
| UNet | - | |
| ResNet-34 | - | |
| ResNet-50 (Robinson et al., 2018) | - | |
| QCResUNet | 1.0 |
3.2 Cardiac MRI segmentation QC task
In the cardiac MRI segmentation QC task, we use the ACDC dataset for training, validation, and testing. The ACDC dataset was partitioned into training, validation, and testing subsets with a ratio of 7:1:2 by following the partitioning rule outlined in (Zhou et al., 2021; Chen et al., 2021; Cao et al., 2022) to avoid overlapping of subjects. This results in a training set consisting of 140 volumes, a validation set with 20 volumes, and a testing set containing 40 volumes. By employing the same four-step approach as in the brain tumor QC task, the original ACDC training, validation, and testing subsets were extended to include 4,900, 640, and 1,280 segmentations. The testing set was then resampled to include 1,011 segmentations. We used the same hyperparameters from the brain tumor segmentation QC task to train the model for the cardiac segmentation QC task. Similar to the brain tumor segmentation QC task, we also reported the mean ( standard deviation) of MAEs and DSC over all subjects for the cardiac segmentation QC task.
| Internal | External | ||||||||
| BraTS [testing] | BraTS-SSA | WUSM | |||||||
| nnUNet+nnFormer | DeepMedic | nnUNet+nnFormer+DeepMedic | nnUNet+nnFormer+DeepMedic | ||||||
| Metric | MAE | MAE | MAE | MAE | |||||
| RCA (Valindria et al., 2017) | NSD DSC | 0.662 0.624 | 0.255 0.165 0.315 0.227 | 0.631 0.660 | 0.272 0.171 0.329 0.215 | 0.690 0.604 | 0.260 0.160 0.302 0.227 | 0.736 0.504 | 0.242 0.158 0.308 0.246 |
| UE-based (Jungo et al., 2020) | NSD DSC | 0.817 0.784 | 0.102 0.095 0.144 0.112 | 0.744 0.762 | 0.113 0.083 0.149 0.114 | 0.627 0.703 | 0.149 0.114 0.165 0.122 | 0.591 0.617 | 0.170 0.119 0.187 0.141 |
| UNet | NSD DSC | 0.939 0.944 | 0.069 0.053 0.077 0.063 | 0.902 0.939 | 0.087 0.054 0.080 0.069 | 0.946 0.931 | 0.061 0.050 0.079 0.074 | 0.903 0.885 | 0.080 0.073 0.102 0.108 |
| ResNet-34 | NSD DSC | 0.944 0.955 | 0.070 0.054 0.072 0.057 | 0.916 0.954 | 0.086 0.065 0.074 0.059 | 0.950 0.943 | 0.067 0.052 0.076 0.068 | 0.894 0.888 | 0.094 0.082 0.103 0.107 |
| ResNet-50 (Robinson et al., 2018) | NSD DSC | 0.943 0.960 | 0.069 0.052 0.070 0.053 | 0.917 0.947 | 0.084 0.061 0.074 0.056 | 0.946 0.941 | 0.066 0.049 0.077 0.068 | 0.895 0.895 | 0.094 0.082 0.103 0.107 |
| QCResUNet | NSD DSC | 0.958∗ 0.968∗ | 0.056 0.043∗ 0.064 0.048∗ | 0.937∗ 0.962∗ | 0.074 0.050∗ 0.062 0.047∗ | 0.954∗ 0.964∗ | 0.057 0.044∗ 0.060 0.049∗ | 0.920∗ 0.912∗ | 0.075 0.062∗ 0.087 0.097∗ |
-
•
∗: ; with a paired t-test to all baseline methods.
| ACDC [testing] | ||||
| NSD | DSC | |||
| MAE | MAE | |||
| RCA | 0.760 | 0.185 0.151 | 0.808 | 0.291 0.136 |
| UE-based | 0.827 | 0.088 0.064 | 0.859 | 0.085 0.063 |
| UNet | 0.884 | 0.071 0.060 | 0.930 | 0.059 0.053 |
| ResNet-34 | 0.891 | 0.073 0.061 | 0.931 | 0.059 0.053 |
| ResNet-50 | 0.883 | 0.076 0.059 | 0.938 | 0.061 0.049 |
| QCResUNet | 0.914∗ | 0.070 0.047∗ | 0.955∗ | 0.057 0.040∗ |
-
•
∗: ; with a paired t-test to all baseline methods.
3.3 Baseline methods
To evaluate the effectiveness of the proposed model, we compared its performance with five baseline models: (i) RCA method (Robinson et al., 2017); (ii) uncertainty estimation (UE) based method (Jungo et al., 2020); and three regression-based methods: (iii) a UNet model (Ronneberger et al., 2015); (iv) a ResNet-34 model (He et al., 2016); and (v) a ResNet-50 (He et al., 2016) model (Robinson et al., 2018).
The selection of the three regression-based methods (UNet, ResNet-34, ResNet-50) as baselines was based on their architectural resemblance to the proposed approach. To ensure a fair comparison, the residual blocks in ResNet-34 and ResNet-50 were identical to those used in the proposed QCResUNet. As the UNet model typically outputs a segmentation mask of the same dimensions as the input, we added an average pooling layer after its final feature map followed by a fully connected layer to enable the prediction of a single DSC value. The RCA and UE-based methods are considered state-of-the-art in the segmentation QC literature.
RCA for brain tumor segmentation QC was implemented using as segmentation method the DeepMedic framework, which was adapted following the protocol described by Valindria et al. (2017). Specifically, we reduced the number of feature maps in each layer by one third compared to the default setting of DeepMedic. We also reduced the feature maps in the last fully connected layer from 150 to 45. We opted for this approach instead of the atlas-based segmentation approach because establishing spatial correspondences between query and reference images in tumorous areas can be challenging due to the high spatial and phenotypic heterogeneity of brain tumors. In contrast, RCA for cardiac MRI segmentation QC was implemented using the atlas-based segmentation method by following the protocols in (Robinson et al., 2019). This is because registration correspondences between query and reference are easier to establish in healthy autonomy. For the selection of the reference data set, we randomly selected 100 samples from the training dataset following a previously well-validated protocol (Robinson et al., 2019).
The implementation of the UE-based method followed the protocol in (Jungo et al., 2020), where the uncertainty map was calibrated by evaluating the uncertainty-error (UE) overlap. The UE overlap measures the overlap between the binarized uncertainty map and the corresponding SEM (see B.1 for details). We chose the UE calibration proposed by Jungo et al. (2020) instead of other calibration methods (Wen et al., 2020; Ashukha et al., 2020; Mehta et al., 2022) because it produces a binary mask by thresholding the uncertainty map. We can directly compare the resulting binary mask to the SEM generated by the proposed QCResUNet. The subject-level DSC was computed by applying a random forest with 102 extracted radiomics features (see B.2).
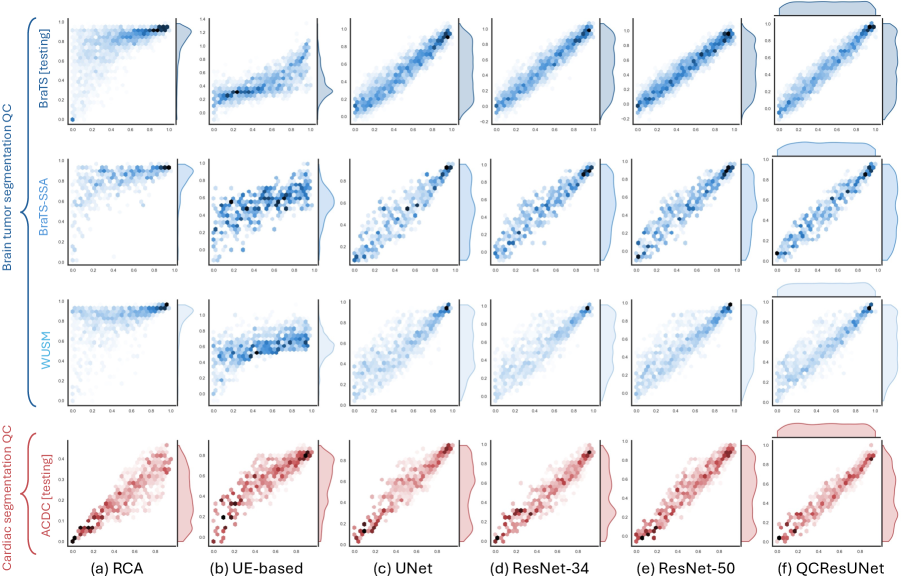

3.4 Evaluation metrics
We evaluated the performance of QC segmentation at both the subject level and the voxel level. At the subject level, we evaluated the precision of the prediction of the segmentation quality metric based on the MAE and the Pearson correlation coefficient . MAE measures the amount of error in the prediction. measures the linear correlation between the predicted DSC/NSD value (DSCpred/NSDpred) and the ground truth DSC/NSD (DSCgt/NSDgt). It ranges from (strongly uncorrelated) to (strongly correlated).
At the voxel level, the performance was evaluated based on the DSC (DSC) between the predicted SEM (SEM) and ground-truth SEM (SEM) for each tissue class . Here, we report the average DSC across all tissue classes. We performed a paired t-test to compare the DSC predicted by the QCResUNet with those predicted by the corresponding baselines. The resulting p-value () was reported as a measure to determine if there was a statistically significant difference between the performance of the proposed method and the baseline methods. The significance level was set a priori to .
3.5 Implementation details
Training procedures were the same for both tasks by using segmentation results were produced by nnUNet, nnFormer, and SegGen. Training was carried out using Adam optimizer (Kingma and Ba, 2014) with a batch size of 4 for 100 epochs. Training was started with an optimal initial learning rate determined by the hyperparameter search and was exponentially decayed by a factor of 0.9 for each epoch until it reached . We applied an weight decay of in all trainable layers. To prevent overfitting, various data augmentations, including random rotation, scaling, mirroring, Gaussian noise, and gamma intensity correction, were applied during training. All model training was performed on NVIDIA Tesla A100 GPUs. By default, we used a mix of single-precision (FP32) and half-precision (FP16) tensors during training to reduce training time and memory consumption. The proposed method was implemented using PyTorch v1.12.1. All the code and pre-trained models developed for this work will be available at https://github.com/sotiraslab/QCResUNet.
4 Results
4.1 Evaluation of subject-level QC performance
4.1.1 Brain tumor MRI segmentation QC task
The proposed model performed well in the subject-level DSC and NSD prediction across all three brain tumor datasets (Table 3). Specifically, on the BraTS internal testing set with segmentations generated by nnUNet and nnFormer, the proposed method achieved a small mean MAE of 0.056 and 0.064 for NSD and DSC prediction, respectively. The predicted NSD and DSC also showed a strong correlation with the corresponding ground truth, achieving Pearson r values of 0.958 and 0.968, respectively. Importantly, the proposed method generalized well for segmentation produced by different methods (i.e., DeepMedic), which had not been used during training, achieving an average MAE of 0.074 and 0.062 for NSD and DSC prediction. Similarly, the predicted NSD and DSC showed a strong correlation with their ground truth with Pearson r values of 0.937 and 0.962, respectively.
Critically, our method also generalized well to completely unseen external BraTS-SSA and WUSM datasets. On the BraTS-SSA dataset, the proposed method achieved an MAE of and for NSD and DSC prediction, respectively. In addition, the Pearson between the predicted segmentation quality measures and their ground-truth values demonstrated a high correlation (NSD ; DSC ). Despite containing MRI scans with varying image quality and tumor characteristics, the proposed method still generalized well to the BraTS-SSA dataset. However, there was a slight drop in performance on the WUSM dataset with a Pearson r of 0.920 and 0.912 for NSD and DSC predictions, respectively. The MAE for the NSD and DSC prediction on the WUSM dataset was 0.075 and 0.087, respectively. We conjectured this might be attributed to domain shift due to differences in data acquisition, preprocessing, and the variability of shape and structures in brain tumors.
Importantly, the proposed method outperformed all baseline methods in NSD and DSC prediction tasks. Compared to the three regression-based QC methods (i.e., UNet, ResNet-34, ResNet-50), QCResUNet improved the second-best method by 0.8% for NSD prediction and 1.5% for DSC prediction in terms of Pearson r values on the BraTS internal testing set. On the external BraTS-SSA and WUSM datasets, QCResUNet outperformed the second-best method by an average of 1.3% and 1.9% in terms of Pearson r, respectively. A paired t-test confirmed that this improvement was statistically significant compared to all three regression-based methods baseline (Table 3). The proposed approach exhibited more evenly distributed DSC prediction errors across different quality levels compared to all the baseline methods (refer to Figure 3 and Figure 4), demonstrating a smaller standard deviation in MAE (see Table 3).
| Brain tumor segmentation QC | Cardiac segmentation QC | ||||
| BraTS [testing] | BraTS-SSA | WUSM | ACDC [testing] | ||
| UE-based (Jungo et al., 2020) | 0.360 0.223 | 0.309 0.106 | 0.283 0.150 | 0.460 0.038 | |
| ablation | QCResUNet w/o attn. | 0.633 0.190 | 0.641 0.052 | 0.618 0.100 | 0.613 0.116 |
| QCResUNet w/o regr. | 0.746 0.164 | 0.685 0.115 | 0.674 0.076 | 0.668 0.091 | |
| QCResUNet | 0.769 0.131∗ | 0.702 0.088∗ | 0.684 0.073∗ | 0.703 0.082∗ | |
-
•
∗: ; with a paired t-test to all baseline methods. QCResUNet w/o attn. refers to QCResUNet without the proposed attention-based SEM aggregation. QCResUNet w/o regr. indicates QCResUNet without performing subject-level QC of DSC and NSD. QCResUNet represents the proposed QCResUNet that utilizes attention-based SEM aggregation and performs both subject-level and voxel-level QC prediction.


The proposed method demonstrated a strong performance gain over the state-of-the-art RCA and UE-based QC methods (Table 3). The performance of the RCA and the UE-based method after hyper-parameter tuning was in line with previous works (Robinson et al., 2017; Jungo et al., 2020). In the internal BraTS testing set, the proposed method improved the average Pearson r of NSD predictions by 48.5% and DSC predictions by 22.1% compared to RCA and UE-based QC, respectively. A similar trend was observed on the external datasets, with the proposed method improving Pearson by an average of and compared to RCA and UE-based QC, respectively. Moreover, the proposed method achieved a significant reduction in the average MAE of predicting NSD and DSC compared to the RCA and UE-based methods for all results viz. internal testing results (, vs. ), BraTS-SSA results (, vs. ), and WUSM results (, vs. ).
4.1.2 Cardiac MRI segmentation QC task
Similar to the brain tumor segmentation QC task, the proposed method achieved a good performance on the cardiac segmentation QC task (see Table 4). Specifically, the proposed method achieved average MAEs of 0.070 and 0.057 for NSD and DSC predictions, respectively. The predicted NSD and DSC demonstrated a strong correlation with the corresponding ground truth, with Pearson r values of 0.914 and 0.955, respectively. The proposed QCResUNet also outperformed all baseline methods in the cardiac segmentation QC task. QCResUNet improved the second-best regression-based method by 2.6% and 1.8% in Pearson r for NSD and DSC predictions, respectively. In addition, QCResUNet significantly outperformed RCA and UE-based QC methods by an average of 10.2% and 10.9%. Lastly, the proposed method achieved a significant reduction in MAEs compared to all baselines (Table 4), aligning well with the ground truth segmentation quality measures (see Figure 3 and Figure 4).
4.2 Evaluation of segmentation error localization
The proposed QCResUNet achieved good voxel-level segmentation error localization in terms of DSC for both brain tumor and cardiac segmentation QC tasks (see Table 5). Specifically, QCResUNet achieved an average DSC of 0.769 on the BraTS internal testing set, 0.702 on the BraTS-SSA dataset, and 0.684 on the WUSM dataset in the brain tumor segmentation QC task. Despite a slight performance drop on external datasets, the results remain high quality. Inter-rater agreement studies in glioma segmentation (Visser et al., 2019) indicate that an overlap measure above 0.7 signifies a good segmentation result. Our out-of-distribution DSC results were on average around 0.7, despite the increased difficulty of the task. In addition, QCResUNet achieved an average DSC of 0.703 in the cardiac segmentation QC task. The detailed distribution of tissue-level DSC (i.e., [DSC, DSC, DSC] in brain tumor segmentation and [DSC, DSC, DSC] in cardiac segmentation) is shown in Figure 5.
Compared to the UE-based QC method, QCResUNet improved error localization significantly by an average of 127.5% in terms of average DSC on the brain tumor segmentation QC task (see Table 5). Similarly, QCResUNet significantly outperformed the UE-based QC method by 52.8% in the cardiac segmentation QC task for segmentation error localization (Table 5).
Visual demonstration. Overall, the proposed QCResUNet achieved reliable localization of tissue-specific segmentation failures at the voxel level for different levels of segmentation quality (see Figure 6(a)). This is a unique feature of the proposed approach as none of the regression-based baseline methods can offer error localization. However, we observed that the performance of segmentation error localization might drop when the query segmentation is of high quality (Figure 6(b)). This performance drop likely arises from the inherent challenge of detecting small errors at the boundaries of high-quality segmentations. However, these cases are less critical to correct, as the query segmentation has already achieved good quality. Therefore, this drop in performance has a minor impact on the potential clinical applicability of the proposed approach.
4.3 Ablation analysis
We performed ablation studies to validate the effectiveness of the proposed multi-task learning strategy and attention-based SEM aggregation mechanism. Although sharing the same network structure for subject-level segmentation quality prediction, QCResUNet outperformed ResNet-34 in both brain tumor and cardiac segmentation tasks (Table 3 and Table 4). In addition, we found that the performance of voxel-level segmentation error prediction deteriorated when removing the subject-level prediction task (see Table 5). This demonstrates the effectiveness of the proposed multi-task learning framework for segmentation QC tasks.
In the voxel-level segmentation error localization task, incorporating the proposed attention-based SEM aggregation mechanism improved performance by an average of 13.9% for the brain tumor segmentation QC task and 14.7% for the cardiac segmentation QC task. We conjectured the improvement introduced by the proposed attention-based SEM aggregation could be attributed to the fact that the features in the last layer of the decoder contribute differently to the prediction of each tissue class error segmentation mask.
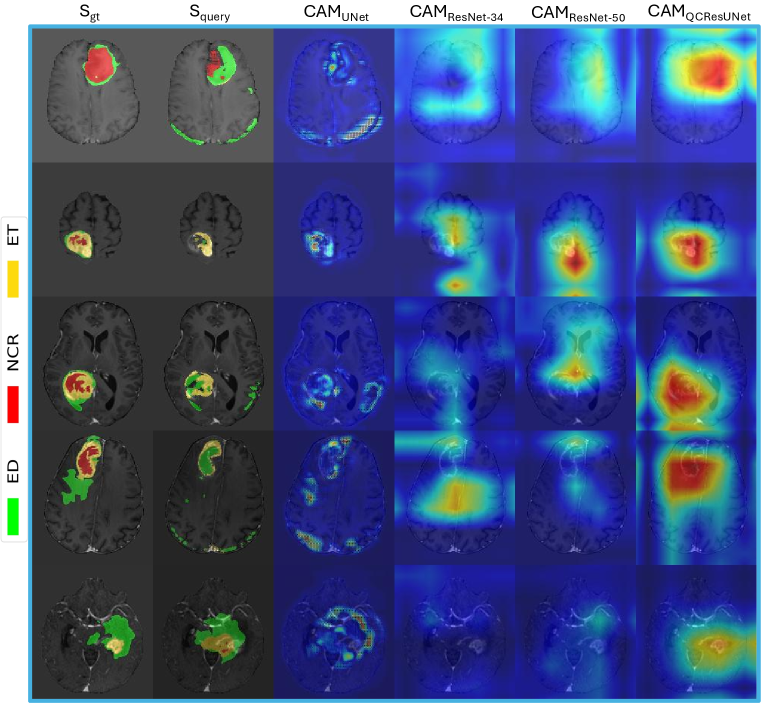
4.4 Explainability analysis
We hypothesized that the performance gain of the proposed QCResUnet compared to the baselines of UNet, ResNet-34, and ResNet-50 is due to joint training under both subject-level and voxel-level supervision. One potential explanation could be that the joint training allowed the proposed QCResUNet to effectively localize on segmentation errors, as suggested by the attention maps produced by Gradient-weighted Class Activation Mapping (Grad-CAM) (Selvaraju et al., 2017) (see Figure 7). We observed that the CAM obtained from ResNet-34 and ResNet-50 did not focus on the areas of segmentation errors. Although ResNet-50 had more parameters and depth, it did not demonstrate significant improvement over ResNet-34, indicating that these factors did not improve the QC performance. In contrast, the UNet CAM showed a coarse localization on the difference between the ground truth and the predicted segmentation. In contrast to the baseline methods, the QCResUNet CAM was well localized on segmentation errors.
5 Discussion and conclusions
In this work, we proposed QCResUNet, a novel 3D CNN architecture designed for automated QC of multi-class tissue segmentation in MRI scans. To the best of our knowledge, this is the first study to provide reliable simultaneous subject-level segmentation quality predictions and voxel-level identification of segmentation errors for different tissue classes. The results suggest that the proposed method is a promising approach for large-scale automated segmentation QC and for guiding clinicians’ feedback for refining segmentation results.
A key feature of the proposed method is the multi-task objective. This enabled the proposed method to focus on regions where errors have occurred, leading to improved performance. This is supported by the following observations. First, we observed that the CAM of the QCResUNet encoder focused more on the regions where the segmentation error occurred compared to ResNet-34 and ResNet-50 (refer to Figure 7). Second, we observed that the supervision from the segmentation error prediction task can in turn guide the DSC and NSD prediction task to prioritize these error-prone regions. As suggested by the CAM, ResNet-34 and ResNet-50 achieved more accurate DSC and NSD prediction than the UNet, while the UNet performed better in segmentation error localization. The possible reason behind this is that the final average pooling layer in the UNet treats each element in the last feature map equally while ignoring the actual size of tumors. In contrast, the average pooling in the embedded feature space in ResNet-based methods operates on the abstracted quality feature maps to preserve information, which resulted in better predictive performance. In contrast, the joint optimization of both subject-level and voxel-level predictions by the proposed QCResUNet allows it to combine the advantages of both ResNet-based models and UNet. As a consequence, QCResUNet can simultaneously localize segmentation errors and assess the overall quality of the segmentation.
Importantly, the proposed method exhibited high generalizability when applied to unseen data, surpassing other state-of-the-art segmentation QC methods. This was particularly true for the RCA and UE-based methods in the brain tumor segmentation QC task, which exhibited poor performance when assessing the quality of segmentation results obtained using segmentation methods different than the ones used to generate training data. The poor generalizability of the RCA method was mainly due to the inherent difficulty of obtaining a representative reference dataset for brain tumor segmentation, which is subject to significant variability. Such significant variability may violate the underlying assumption of the RCA method that there is at least one sample in the reference dataset that can be successfully segmented given a query image-segmentation pair (Robinson et al., 2017; Valindria et al., 2017), which is only valid when dealing with healthy anatomies. In the case of the cardiac segmentation QC task, which involves healthy anatomy, RCA methods achieved better performance compared to the brain tumor case when implemented with the atlas-based segmentation method. Similar to RCA, the UE-based QC did not perform well on subject-level quality prediction as well as localizing segmentation error in the brain tumor case. This may be attributed to the fact that there is inherent variability in uncertainty maps produced by various segmentation methods on datasets with different image quality, tumor characteristics, etc. While in the cardiac segmentation QC task, which involves healthy anatomy and less variability, the UE-based showed better performance. Additionally, as consistent with findings in Jungo et al. (2020), we found that the UE-based method offers limited segmentation error localization. This limitation further hinders its ability to generalize effectively. Furthermore, the UE-based method can only be used to assess segmentations obtained from deep learning models. Unless the deep learning segmentation method directly outputs an estimate of voxel-wise uncertainty, test time estimation of uncertainty (e.g., using MCDropout (Gal and Ghahramani, 2016)) requires access to the model architecture and weights, which may not be possible for models deployed in clinical settings.
The proposed work is not without limitations. Firstly, though the proposed method has better generalizability than other segmentation QC methods, it may be affected by domain shift issues, which are common to all deep learning methods. As a consequence, translating the proposed to clinical practice will require monitoring of each performance to ensure reliable results. In addition, techniques that can enhance the robustness and generalizability of the proposed method to domain shifts (Ganin et al., 2016; Carlucci et al., 2019) are worth further investigating in future work. Second, a key requirement of the proposed method is the availability of all four modalities. Although handling missing modalities enhances the applicability of the proposed method in diverse clinical settings, it is a non-trivial problem. The integration of existing techniques for handling missing modalities (Dorent et al., 2019; Shen and Gao, 2019; Wang et al., 2023) into the proposed QCResUNet will increase significantly the computational burden for training. This is due to the fact that these methods necessitate separate sets of encoders and decoders for each input modality, resulting in a computational load that is at least quadruple that of the current QCResUNet. In our future work, we will explore efficient strategies to allow the model to handle missing modalities. Third, the proposed method was only validated on segmentation tasks involving a single object. Although we have shown that the proposed method generalized well across different segmentation tasks that include multiple tissues, more experiments are needed to evaluate how this method performs in the presence of multiple objects (e.g., multiple lesions). Lastly, this work focused on predicting DSC and NSD as metrics to appropriately summarize segmentation quality. However, it is important to recognize that these metrics have limitations and may not be suitable for all applications. While we demonstrated the proposed method’s ability to handle multiple metrics, additional research will be required to tailor the method for predicting the most appropriate metric for specific tasks (Maier-Hein et al., 2024).
To conclude, we developed QCResUNet for the automated brain tumor and cardiac segmentation QC. Our proposed method is able to reliably assess segmentation quality at the subject level, while at the same time accurately identifying tissue-specific segmentation errors at the voxel level. Through multi-task learning under subject-level and voxel-level supervision, we achieved strong performance in both prediction tasks. Training the network on a large-scale dataset, which comprised segmentation results from various methods and at different levels of quality, allowed the proposed method to generalize well to unseen data. A key characteristic of the proposed method is that it is agnostic to the method used to generate the segmentation. This makes it versatile for evaluating the quality of segmentation results generated by different methods. A unique characteristic of the proposed method is its ability to accurately pinpoint the location of tissue-specific segmentation errors, thus potentially facilitating the integration of human input for refining automatically generated segmentations in clinical settings. This, in turn, has the potential to enhance clinical workflows.
References
- Adewole et al. (2023) Adewole, M., Rudie, J.D., Gbdamosi, A., Toyobo, O., Raymond, C., Zhang, D., Omidiji, O., Akinola, R., Suwaid, M.A., Emegoakor, A., et al., 2023. The brain tumor segmentation (brats) challenge 2023: Glioma segmentation in sub-saharan africa patient population (brats-africa). ArXiv .
- Ahn et al. (2019) Ahn, J., Cho, S., Kwak, S., 2019. Weakly supervised learning of instance segmentation with inter-pixel relations, in: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp. 2209–2218.
- Albà et al. (2018) Albà, X., Lekadir, K., Pereañez, M., Medrano-Gracia, P., Young, A.A., Frangi, A.F., 2018. Automatic initialization and quality control of large-scale cardiac mri segmentations. Medical Image Analysis 43, 129–141.
- Ashukha et al. (2020) Ashukha, A., Lyzhov, A., Molchanov, D., Vetrov, D., 2020. Pitfalls of in-domain uncertainty estimation and ensembling in deep learning. arXiv preprint arXiv:2002.06470 .
- Bai et al. (2018) Bai, W., Sinclair, M., Tarroni, G., Oktay, O., Rajchl, M., Vaillant, G., Lee, A.M., Aung, N., Lukaschuk, E., Sanghvi, M.M., et al., 2018. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. Journal of Cardiovascular Magnetic Resonance 20, 1–12.
- Baid et al. (2021) Baid, U., Ghodasara, S., Mohan, S., Bilello, M., Calabrese, E., Colak, E., Farahani, K., Kalpathy-Cramer, J., Kitamura, F.C., Pati, S., Prevedello, L.M., Rudie, J.D., Sako, C., Shinohara, R.T., Bergquist, T., Chai, R., Eddy, J., Elliott, J., Reade, W., Schaffter, T., Yu, T., Zheng, J., Moawad, A.W., Coelho, L.O., McDonnell, O., Miller, E., Moron, F.E., Oswood, M.C., Shih, R.Y., Siakallis, L., Bronstein, Y., Mason, J.R., Miller, A.F., Choudhary, G., Agarwal, A., Besada, C.H., Derakhshan, J.J., Diogo, M.C., Do-Dai, D.D., Farage, L., Go, J.L., Hadi, M., Hill, V.B., Iv, M., Joyner, D., Lincoln, C., Lotan, E., Miyakoshi, A., Sanchez-Montano, M., Nath, J., Nguyen, X.V., Nicolas-Jilwan, M., Jimenez, J.O., Ozturk, K., Petrovic, B.D., Shah, C., Shah, L.M., Sharma, M., Simsek, O., Singh, A.K., Soman, S., Statsevych, V., Weinberg, B.D., Young, R.J., Ikuta, I., Agarwal, A.K., Cambron, S.C., Silbergleit, R., Dusoi, A., Postma, A.A., Letourneau-Guillon, L., Perez-Carrillo, G.J.G., Saha, A., Soni, N., Zaharchuk, G., Zohrabian, V.M., Chen, Y., Cekic, M.M., Rahman, A., Small, J.E., Sethi, V., Davatzikos, C., Mongan, J., Hess, C., Cha, S., Villanueva-Meyer, J., Freymann, J.B., Kirby, J.S., Wiestler, B., Crivellaro, P., Colen, R.R., Kotrotsou, A., Marcus, D., Milchenko, M., Nazeri, A., Fathallah-Shaykh, H., Wiest, R., Jakab, A., Weber, M.A., Mahajan, A., Menze, B., Flanders, A.E., Bakas, S., 2021. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification .
- Bernard et al. (2018) Bernard, O., Lalande, A., Zotti, C., Cervenansky, F., Yang, X., Heng, P.A., Cetin, I., Lekadir, K., Camara, O., Ballester, M.A.G., et al., 2018. Deep learning techniques for automatic mri cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE transactions on medical imaging 37, 2514–2525.
- Cao et al. (2022) Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., Wang, M., 2022. Swin-unet: Unet-like pure transformer for medical image segmentation, in: European conference on computer vision, Springer. pp. 205–218.
- Carlucci et al. (2019) Carlucci, F.M., D’Innocente, A., Bucci, S., Caputo, B., Tommasi, T., 2019. Domain generalization by solving jigsaw puzzles, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR).
- Chakrabarty et al. (2022) Chakrabarty, S., Abidi, S.A., Mousa, M., Mokkarala, M., Hren, I., Yadav, D., Kelsey, M., LaMontagne, P., Wood, J., Adams, M., Su, Y., Thorpe, S., Chung, C., Sotiras, A., Marcus, D.S., 2022. Integrative imaging informatics for cancer research: Workflow automation for neuro-oncology (i3cr-wano).
- Chen et al. (2021) Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., Lu, L., Yuille, A.L., Zhou, Y., 2021. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 .
- Dorent et al. (2019) Dorent, R., Joutard, S., Modat, M., Ourselin, S., Vercauteren, T., 2019. Hetero-modal variational encoder-decoder for joint modality completion and segmentation, in: Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part II 22, Springer. pp. 74–82.
- Drozdzal et al. (2016) Drozdzal, M., Vorontsov, E., Chartrand, G., Kadoury, S., Pal, C., 2016. The importance of skip connections in biomedical image segmentation, in: International Workshop on Deep Learning in Medical Image Analysis, International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis, Springer. pp. 179–187.
- Gal and Ghahramani (2016) Gal, Y., Ghahramani, Z., 2016. Dropout as a bayesian approximation: Representing model uncertainty in deep learning , 1050–1059.
- Ganin et al. (2016) Ganin, Y., Ustinova, E., Ajakan, H., Germain, P., Larochelle, H., Laviolette, F., March, M., Lempitsky, V., 2016. Domain-adversarial training of neural networks. Journal of machine learning research 17, 1–35.
- Garcia-Garcia et al. (2017) Garcia-Garcia, A., Orts-Escolano, S., Oprea, S., Villena-Martinez, V., Garcia-Rodriguez, J., 2017. A review on deep learning techniques applied to semantic segmentation. arXiv preprint arXiv:1704.06857 .
- Grady et al. (2012) Grady, L., Singh, V., Kohlberger, T., Alvino, C., Bahlmann, C., 2012. Automatic segmentation of unknown objects, with application to baggage security , 430–444.
- He et al. (2016) He, K., Zhang, X., Ren, S., Sun, J., 2016. Deep residual learning for image recognition , 770–778.
- Ioffe and Szegedy (2015) Ioffe, S., Szegedy, C., 2015. Batch normalization: Accelerating deep network training by reducing internal covariate shift. International conference on machine learning , 448–456.
- Isensee et al. (2018a) Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H., 2018a. Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge , 287–297.
- Isensee et al. (2018b) Isensee, F., Petersen, J., Klein, A., Zimmerer, D., Jaeger, P.F., Kohl, S., Wasserthal, J., Koehler, G., Norajitra, T., Wirkert, S., Maier-Hein, K.H., 2018b. nnu-net: Self-adapting framework for u-net-based medical image segmentation .
- Jungo et al. (2020) Jungo, A., Balsiger, F., Reyes, M., 2020. Analyzing the quality and challenges of uncertainty estimations for brain tumor segmentation. Frontiers in neuroscience , 282.
- Kamnitsas et al. (2017) Kamnitsas, K., Ledig, C., Newcombe, V.F., Simpson, J.P., Kane, A.D., Menon, D.K., Rueckert, D., Glocker, B., 2017. Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Medical Image Analysis 36, 61–78.
- Khened et al. (2019) Khened, M., Kollerathu, V.A., Krishnamurthi, G., 2019. Fully convolutional multi-scale residual densenets for cardiac segmentation and automated cardiac diagnosis using ensemble of classifiers. Medical image analysis 51, 21–45.
- Kingma and Ba (2014) Kingma, D.P., Ba, J., 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 .
- Kofler et al. (2022) Kofler, F., Ezhov, I., Fidon, L., Horvath, I., de la Rosa, E., LaMaster, J., Li, H., Finck, T., Shit, S., Paetzold, J., et al., 2022. Deep quality estimation: Creating surrogate models for human quality ratings. arXiv preprint arXiv:2205.10355 .
- Kohlberger et al. (2012) Kohlberger, T., Singh, V., Alvino, C., Bahlmann, C., Grady, L., 2012. Evaluating segmentation error without ground truth, in: Ayache, N., Delingette, H., Golland, P., Mori, K. (Eds.), Medical Image Computing and Computer-Assisted Intervention – MICCAI 2012, Springer Berlin Heidelberg, Berlin, Heidelberg. pp. 528–536.
- Li et al. (2022) Li, K., Yu, L., Heng, P.A., 2022. Towards reliable cardiac image segmentation: Assessing image-level and pixel-level segmentation quality via self-reflective references. Medical Image Analysis 78, 102426.
- Li et al. (2018) Li, L., Jamieson, K., DeSalvo, G., Rostamizadeh, A., Talwalkar, A., 2018. Hyperband: A novel bandit-based approach to hyperparameter optimization. Journal of Machine Learning Research 18, 1–52.
- Li et al. (2020) Li, L., Jamieson, K., Rostamizadeh, A., Gonina, E., Ben-Tzur, J., Hardt, M., Recht, B., Talwalkar, A., 2020. A system for massively parallel hyperparameter tuning. Proceedings of Machine Learning and Systems 2, 230–246.
- Liaw et al. (2018) Liaw, R., Liang, E., Nishihara, R., Moritz, P., Gonzalez, J.E., Stoica, I., 2018. Tune: A research platform for distributed model selection and training. arXiv preprint arXiv:1807.05118 .
- Litjens et al. (2017) Litjens, G., Kooi, T., Bejnordi, B.E., Setio, A.A.A., Ciompi, F., Ghafoorian, M., Van Der Laak, J.A., Van Ginneken, B., Sánchez, C.I., 2017. A survey on deep learning in medical image analysis. Medical image analysis 42, 60–88.
- Maas et al. (2013) Maas, A.L., Hannun, A.Y., Ng, A.Y., et al., 2013. Rectifier nonlinearities improve neural network acoustic models, in: Proc. icml, Atlanta, Georgia, USA. p. 3.
- Maier-Hein et al. (2024) Maier-Hein, L., Reinke, A., Godau, P., Tizabi, M.D., Buettner, F., Christodoulou, E., Glocker, B., Isensee, F., Kleesiek, J., Kozubek, M., et al., 2024. Metrics reloaded: recommendations for image analysis validation. Nature methods , 1–18.
- Mehta et al. (2022) Mehta, R., Filos, A., Baid, U., Sako, C., McKinley, R., Rebsamen, M., Dätwyler, K., Meier, R., Radojewski, P., Murugesan, G.K., et al., 2022. Qu-brats: Miccai brats 2020 challenge on quantifying uncertainty in brain tumor segmentation-analysis of ranking scores and benchmarking results. The journal of machine learning for biomedical imaging 2022.
- Ng et al. (2020) Ng, M., Guo, F., Biswas, L., Petersen, S.E., Piechnik, S.K., Neubauer, S., Wright, G., 2020. Estimating uncertainty in neural networks for cardiac mri segmentation: a benchmark study. arXiv preprint arXiv:2012.15772 .
- Ng et al. (2018) Ng, M., Guo, F., Biswas, L., Wright, G.A., 2018. Estimating uncertainty in neural networks for segmentation quality control , 3–6.
- Qi et al. (2020) Qi, J., Du, J., Siniscalchi, M., Ma, X., Lee, C.H., 2020. On mean absolute error for deep neural network based vector-to-vector regression. IEEE Signal Processing Letters PP.
- Qiu et al. (2023) Qiu, P., Chakrabarty, S., Nguyen, P., Ghosh, S.S., Sotiras, A., 2023. Qcresunet: Joint subject-level and voxel-level prediction of segmentation quality, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 173–182.
- Robinson et al. (2018) Robinson, R., Oktay, O., Bai, W., Valindria, V.V., Sanghvi, M.M., Aung, N., Paiva, J.M., Zemrak, F., Fung, K., Lukaschuk, E., et al., 2018. Real-time prediction of segmentation quality , 578–585.
- Robinson et al. (2019) Robinson, R., Valindria, V.V., Bai, W., Oktay, O., Kainz, B., Suzuki, H., Sanghvi, M.M., Aung, N., Paiva, J.M., Zemrak, F., et al., 2019. Automated quality control in image segmentation: application to the uk biobank cardiovascular magnetic resonance imaging study. Journal of Cardiovascular Magnetic Resonance 21, 1–14.
- Robinson et al. (2017) Robinson, R., Valindria, V.V., Bai, W., Suzuki, H., Matthews, P.M., Page, C., Rueckert, D., Glocker, B., 2017. Automatic quality control of cardiac mri segmentation in large-scale population imaging , 720–727.
- Rohlfing et al. (2010) Rohlfing, T., Zahr, N.M., Sullivan, E.V., Pfefferbaum, A., 2010. The sri24 multichannel atlas of normal adult human brain structure. Human brain mapping 31, 798–819.
- Ronneberger et al. (2015) Ronneberger, O., Fischer, P., Brox, T., 2015. U-net: Convolutional networks for biomedical image segmentation , 234–241.
- Roy et al. (2018) Roy, A.G., Conjeti, S., Navab, N., Wachinger, C., 2018. Inherent brain segmentation quality control from fully convnet monte carlo sampling , 664–672.
- Sander et al. (2020) Sander, J., de Vos, B.D., Išgum, I., 2020. Automatic segmentation with detection of local segmentation failures in cardiac mri. Scientific Reports 10, 1–19.
- Selvaraju et al. (2017) Selvaraju, R.R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., Batra, D., 2017. Grad-cam: Visual explanations from deep networks via gradient-based localization, in: Proceedings of the IEEE international conference on computer vision, pp. 618–626.
- Shen and Gao (2019) Shen, Y., Gao, M., 2019. Brain tumor segmentation on mri with missing modalities, in: Information Processing in Medical Imaging: 26th International Conference, IPMI 2019, Hong Kong, China, June 2–7, 2019, Proceedings 26, Springer. pp. 417–428.
- Tompson et al. (2015) Tompson, J., Goroshin, R., Jain, A., LeCun, Y., Bregler, C., 2015. Efficient object localization using convolutional networks. Proceedings of the IEEE conference on computer vision and pattern recognition , 648–656.
- Tran (2016) Tran, P.V., 2016. A fully convolutional neural network for cardiac segmentation in short-axis mri. arXiv preprint arXiv:1604.00494 .
- Ulyanov et al. (2016) Ulyanov, D., Vedaldi, A., Lempitsky, V., 2016. Instance normalization: The missing ingredient for fast stylization. arXiv preprint arXiv:1607.08022 .
- Valindria et al. (2017) Valindria, V.V., Lavdas, I., Bai, W., Kamnitsas, K., Aboagye, E.O., Rockall, A.G., Rueckert, D., Glocker, B., 2017. Reverse classification accuracy: predicting segmentation performance in the absence of ground truth. IEEE transactions on medical imaging 36, 1597–1606.
- Visser et al. (2019) Visser, M., Müller, D., van Duijn, R., Smits, M., Verburg, N., Hendriks, E., Nabuurs, R., Bot, J., Eijgelaar, R., Witte, M., et al., 2019. Inter-rater agreement in glioma segmentations on longitudinal mri. NeuroImage: Clinical 22, 101727.
- Wang et al. (2023) Wang, H., Chen, Y., Ma, C., Avery, J., Hull, L., Carneiro, G., 2023. Multi-modal learning with missing modality via shared-specific feature modelling, in: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 15878–15887.
- Wang et al. (2020a) Wang, Q., Wu, B., Zhu, P., Li, P., Zuo, W., Hu, Q., 2020a. Eca-net: Efficient channel attention for deep convolutional neural networks, in: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp. 11534–11542.
- Wang et al. (2020b) Wang, S., Tarroni, G., Qin, C., Mo, Y., Dai, C., Chen, C., Glocker, B., Guo, Y., Rueckert, D., Bai, W., 2020b. Deep generative model-based quality control for cardiac mri segmentation , 88–97.
- Wen et al. (2020) Wen, Y., Tran, D., Ba, J., 2020. Batchensemble: an alternative approach to efficient ensemble and lifelong learning. arXiv preprint arXiv:2002.06715 .
- Zhou et al. (2021) Zhou, H.Y., Guo, J., Zhang, Y., Yu, L., Wang, L., Yu, Y., 2021. nnformer: Interleaved transformer for volumetric segmentation. arXiv preprint arXiv:2109.03201 .

Appendix A Subject-level multi-class DSC
We calculated the subject-level DSC across all tissue classes using the standard definition:
| (1) |
where TP, FP, and FN denote true positive, false positive, and false negative, respectively. For a certain location in the multi-class query segmentation mask (), the predicted foreground label is considered a true positive if and only if it matches the corresponding ground truth label. Similarly, the predicted background label is considered a false positive if and only if it matches the true background. Otherwise, if a background label is incorrectly predicted as a foreground label, it is counted as a false positive.
The rationale for selecting this multi-class DSC as a subject-level quality measure is to provide a single value that summarizes the overall quality of the segmentation. This is beneficial for streamlined image-driven analysis, as it allows us to filter out low-quality segmentation cases using a single threshold.
Appendix B Uncertainty estimation-based QC
The uncertainty estimation based QC was implemented using the Monte Carlo Dropout (MCDropout) approach (Gal and Ghahramani, 2016), given its success and popularity. MCDropout was applied to the main convolution blocks in the nnUNet and DeepMedic as well as to the self-attention block in the nnFormer with a drop rate , following the protocol in (Jungo et al., 2020). After running the inference of the model times, the uncertainty map of a segmentation can be computed as
| (2) |
where and denote the voxel index and the tumor tissue types, while denotes the probability map obtained from the softmax layer of a trained segmentation model. was set empirically to 50 to strike a balance between the number of Monte-Carlo samples we draw and computational complexity.
B.1 Uncertainty-Error Overlap
We used the uncertainty-error (UE) overlap proposed by Jungo et al. (2020) to calibrate the uncertainty map. The UE overlap measures the overlap between the uncertainty map (UMap) and the ground-truth segmentation error map (SEMgt) following the DSC formula:
where denotes the cardinality of the set of voxels of each binary map. To compute the UE overlap, the uncertainty map UMap needs to be thresholded into a binary mask. Following the protocol in Jungo et al. (2020), the optimal threshold value was determined by examining different thresholds in the validation set in increments of 0.05, ranging from 0.05 to 0.95. The optimal threshold values for different datasets are shown in Table 1.
| Brain tumor segmentation QC | Cardiac segmentation QC | |||
| Threshold | BraTS [testing] | BraTS-SSA | WUSM | ACDC [testing] |
| nnUNet | [0.10, 0.10, 0.10] | [0.10, 0.10, 0.15] | [0.15, 0.10, 0.15] | [0.10, 0.10, 0.10] |
| nnFormer | [0.10, 0.10, 0.30] | [0.30, 0.35, 0.35] | [0.40, 0.30, 0.40] | [0.10, 0.15, 0.15] |
| DeepMedic | [0.05, 0.05, 0.05] | [0.25, 0.25, 0.20] | [0.05, 0.05, 0.10] | - |
B.2 Subject-level DSC prediction
Following Jungo et al. (2020), the voxel-level uncertainty map was aggregated to subject-level Dice Similarity Coefficient (DSC) and Normalized Surface Dice (NSD) predictions by using a two-step approach. First, 102 radiomics features were automatically extracted from the uncertainty map using the PyRadiomics111https://pyradiomics.readthedocs.io package. Second, we trained a random forest regressor using scikit-learn222https://scikit-learn.org/stable/ package in the training dataset to predict the subject-level DSC and NSD.
Appendix C Hyper-parameter tuning results
The hyperparameter tuning was carried out using the Raytune333https://docs.ray.io/en/latest/index.html package. Specifically, we utilized a Bandit-based approach for efficient resource allocations that allowed us to dedicate more resources to the more promising hyperparameter combinations (Li et al., 2018, 2020). The learning curve for Raytune hyperparameter tuning in the BraTS validation set is shown in Figure S1. The optimal hyperparameter combinations determined by Raytune can be found in Table 2.
Appendix D Voxel-level multi-class segmentation error mask visualization
Here, we provide the details on how to obtain the visualization in Figure 6. The voxel-level SEM prediction from the proposed QCResUNet is a set of binary masks (e.g., in the brain tumor segmentation task, these masks delineate the whole tumor (WT), tumor core (TC), and enhancing tumor (ET), respectively). For the purpose of visualization, we combined the binary WT, TC, and ET masks into a multi-class mask consisting of ET, NCR and ED classes. Specifically, the ED class was constructed by those voxels whose values were one in the WT binary mask and zero in the TC binary mask. The NCR class was constructed by those voxels whose values were one in the TC binary mask and zero in the ET binary mask. The ET class was constructed by those voxels whose values were one in the ET binary mask.