MadDog:
A Web-based System for Acronym Identification and Disambiguation
Abstract
Acronyms and abbreviations are the short-form of longer phrases and they are ubiquitously employed in various types of writing. Despite their usefulness to save space in writing and reader’s time in reading, they also provide challenges for understanding the text especially if the acronym is not defined in the text or if it is used far from its definition in long texts. To alleviate this issue, there are considerable efforts both from the research community and software developers to build systems for identifying acronyms and finding their correct meanings in the text. However, none of the existing works provide a unified solution capable of processing acronyms in various domains and to be publicly available. Thus, we provide the first web-based acronym identification and disambiguation system which can process acronyms from various domains including scientific, biomedical, and general domains. The web-based system is publicly available at http://iq.cs.uoregon.edu:5000 and a demo video is available at https://youtu.be/IkSh7LqI42M. The system source code is also available at https://github.com/amirveyseh/MadDog.
1 Introduction
Textual contents such as books, articles, reports, and web-blogs in various domains are replete with phrases that are commonly used by people in that field. In order to save space in text writing and also facilitate communication among people who are already familiar with these phrases, the shorthanded form of long phrases, known as acronyms and abbreviations, are frequently used. However, the use of acronyms could also introduce challenges to understand the text, especially for newcomers. More specifically, two types of challenges might hinder reading text with acronyms: 1) In long documents, e.g., a book chapter, an acronym might be defined somewhere in the text and used several times throughout the document. For someone who is not familiar with the definition of the acronym and interested in reading a part of the document, it might be time-consuming to find the definition of the acronym in the document. To solve this problem, an automatic acronym identification tool is required whose goal is to find all acronyms and their definitions that are locally provided in the same document. 2) Some of the acronyms might not be even defined in the document itself. These acronyms are commonly used by writers in a specific domain. To find the correct meaning of them, a reader must look-up the acronym in a dictionary of acronyms. However, due to the shorter length of acronyms compared to their long-form, multiple phrases might be shortened with the same acronym, thereby, they will be ambiguous. In these cases, a deep understanding of the domain is required to recognize the correct meaning of the acronym among all possible long-forms. To solve this issue, a system capable of disambiguating an acronym based on its context is necessary.
Each of the aforementioned problems, i.e., acronym identification (AI) and acronym disambiguation (AD), has been extensively studied by the research community or software developers. One of the methods which are widely used in acronym identification research is proposed by Schwartz and Hearst (2002). This is a rule-based model that utilizes character-match between acronym letters and their context to find the acronym and its long-form in text. Later, some feature-based models have been also used for acronym identification Kuo et al. (2009); Liu et al. (2017). In addition, some of the existing software employs regular expressions for acronym identification in the biomedical domain Gooch (2011). Acronym disambiguation is also approached with feature-based models Wang et al. (2016) or more advanced deep learning methods Wu et al. (2015); Ciosici et al. (2019). The majority of deep models employ word embeddings to compute the similarity between the candidate long-form and the acronym context. In addition to the existing research for AD, there is some web-based software that employ dictionary look-up to expand an acronym to its long-form ABBREX (2018). Note that the methods based on dictionary look-up are not able to disambiguate the acronym if it has multiple meanings.
Despite the progress made on the AI and AD task in the last two decades, there are some limitations in the prior works that prevent achieving a functional system to be used in practice. More specifically, considering the research on the AD task, all of the prior works employ a small-size dataset covering a few hundred to a few thousand long-forms in a specific domain. Therefore, the models trained in these works are not capable to expand all acronyms of a domain or acronyms in other domains other than the one used in the training set. Although in the recent work Wen et al. (2020), authors proposed a big dataset for acronym disambiguation in the medical domain with more than 14 million samples, it is still limited to a specific domain (i.e., medical domain). Another limitation in prior works is that they do not provide a unified system capable of performing both tasks in various domains and to be publicly available. To our knowledge, the only exiting web-based system for AI and AD is proposed by Ciosici and Assent (2018). For acronym identification, this system employs the rule-based model introduced by Schwartz and Hearst (2002). To handle corner cases, they add extra rules in addition to Schwartz’s rules in their system. Unfortunately, they do not provide detailed information about these corner cases and extra rules or any evaluation to assess the performance of the model. For acronym disambiguation, they resort to a statistical model in which a pre-computed vector representation for each candidate long-form is employed to compute the similarity between candidate long-form with the context of the ambiguous acronym represented using another vector. However, there are two limitations with this approach: first, the pre-computed long-form vectors are obtained via only Wikipedia, thus limiting this system to the general domain and incapable of disambiguating acronyms in other domains such as scientific papers or biomedical texts; Second, the AD model based on the pre-computed vectors is a statistical model and is not benefiting from the advanced deep architectures, thereby it might have inferior performance compared to a deep AD model.
To address the shortcomings and limitations of the prior research works or systems for AI and AD, in this work, we introduce a web-based system for acronym identification and disambiguation that is capable of recognizing and expanding acronyms in multiple domains including general (e.g., Wikipedia articles), scientific (e.g., computer science papers), biomedical (e.g., Medline abstracts), or financial (e.g., financial discussions in Reddit). More specifically, we first propose a rule-based model for acronym identification by extending the set of rules proposed by Schwartz and Hearst (2002). We empirically show that the proposed model outperforms both the previous rule-based model and also the existing state-of-the-art deep learning models for acronym identification on the recent benchmark dataset SciAI Veyseh et al. (2020). Next, we use a large dataset created from corpora in various domains for acronym disambiguation to train a deep model for this task. Specifically, we employ a sequential deep model to encode the context of the ambiguous acronym and solve the AD task using a feed-forward multi-class classifier. We also evaluate the performance of the proposed acronym disambiguation model on the recent benchmark dataset SciAD Veyseh et al. (2020).
To summarize, our contributions are:
-
•
The first web-based multi-domain acronym identification and disambiguation system
-
•
Extensive evaluation of the proposed model on the two benchmark datasets SciAI and SciAD
2 System Description
The proposed system is a web-based system consisting of two major components: 1) Acronym Identification which consists of a set of prioritized rules to recognize the mentions of acronyms and their long-forms in the text; 2) Acronym Expansion which involves a dictionary look-up to expand acronyms with only one possible long-form and a pre-trained deep learning model to predict the long-form of an ambiguous acronym using its context. The system takes as input a piece of text and returns the text with highlighted acronyms in which the user can click on the acronyms and their long-form will be shown in a pop-up window. The acronym glossary extracted from the text is also shown at the end of the text. Note that users can also enable/disable the acronym expansion component. This section studies the details of the aforementioned components.
2.1 Acronym Identification
Acronym Identification aims to find the mentions of acronyms and their long-forms in text. This is the first stage in the proposed system to identify the acronyms and their immediate definitions. Generally, this task is modeled as a sequence labeling problem. In our system, we employ a rule-based model to extract acronyms and their meanings from a given text. In particular, the proposed AI model is a collection of rules mainly inspired by the rule introduced in Schwartz and Hearst (2002). More specifically, the following rules are employed in the proposed AI model:
-
•
Acronym Detector: This rule identifies all acronyms in text, regardless of having an immediate definition or not. Specifically, all words that at least 60% of their characters are upper-cased letters and the number of their characters is between 2 and 10 are recognized as an acronym (i.e., short-form).
-
•
Bounded Schwartz’s: Similar to Schwartz and Hearst (2002), we look for immediate definitions of detected acronyms if they follow one of the templates long-form (short-form) or short-form (long-form). In particular, considering the first template, we take the words, where is the number of characters in the acronym, that appear immediately before the parentheses as the candidate long-form111Note that we use the same candidate long-form in other rules too. Then, a sub-sequence of the candidate long-form that some of its characters could form the acronym is selected as the long-form. However, despite the original Schwartz’s rule that does not restrict the first and last word of the long-form to be used in the acronym, we enforce this restriction. This modification could fix erroneous long-form detection by Schwartz’s rule. For instance, in the phrase User-guided Social Media Crawling method (USMC), the modified rule identifies the long-form User-guided Social Media Crawling, excluding the leading word method.
-
•
Character Match: While the Bounded Schwartz’ rule could identify the majority of the long-forms, it might also introduce some noisy meanings. For instance, in the phrase Analyzing Avatar Boundary Matching (AABM), the Bounded Schwartz’s rule identifies Avatar Boundary Matching as the long-form of AABM, missing the starting word Analyzing. To solve this issue and increase the model’s accuracy, we also employ a character match rule that assesses if the initials of the words in the candidate long-form could form the acronym. In the given example, it identifies the full phrase Analyzing Avatar Boundary Matching as the long-form. Since this rule is more restricted and it has higher precision than Bounded Schwartz’s rule, in our system, it has a higher priority than the Bounded Schwartz’s rule.
-
•
Initial Capitals: One issue with the proposed Character Matching rule is that if there is a word in the long-form that is not used in the acronym, the rule fails to correctly identify the long-form. For instance, in the phrase Analysis of Avatar Boundary Matching (AABM) the Character Matching rule fails due to the existence of the word of. To mitigate this issue, we propose another high-precision rule, Initial Capitals. In this rule, if the concatenation of the initials of the words of the candidate long-form which are upper-cased could form the acronym, the candidate is selected as the expanded form of the acronym. This rule has the highest priority in our system.
In addition to the mentioned general rules, we also add some other rules to handle the special cases (e.g., acronyms with a hyphen, roman numbers, definitions provided in some templates (e..g, CNN stands for convolution neural network)).
| Acronym | Long-form | Rule |
|---|---|---|
| AABM | Analyzing Avatar Boundary Matching | Character Match |
| ABBREX | Abbreviation Expander | Bounded Schwartz’s |
| AD | acronym disambiguation | Character Match |
| AI | Acronym identification | Character Match |
| BADREX | Biomedical Abbreviations using | Bounded Schwartz’s |
| Dynamic Regular Expressions | ||
| BiLSTM | Bi - directional Long ShortTerm Memory | Bounded Schwartz’s |
| DOG | Diverse acrOnym Glossary | Bounded Schwartz’s |
| MAD | Massive Acronym Disambiguation | Capital Initials |
| MF | most frequent | Character Match |
| USMC | User - guided Social Media Crawling | Capital Initials |
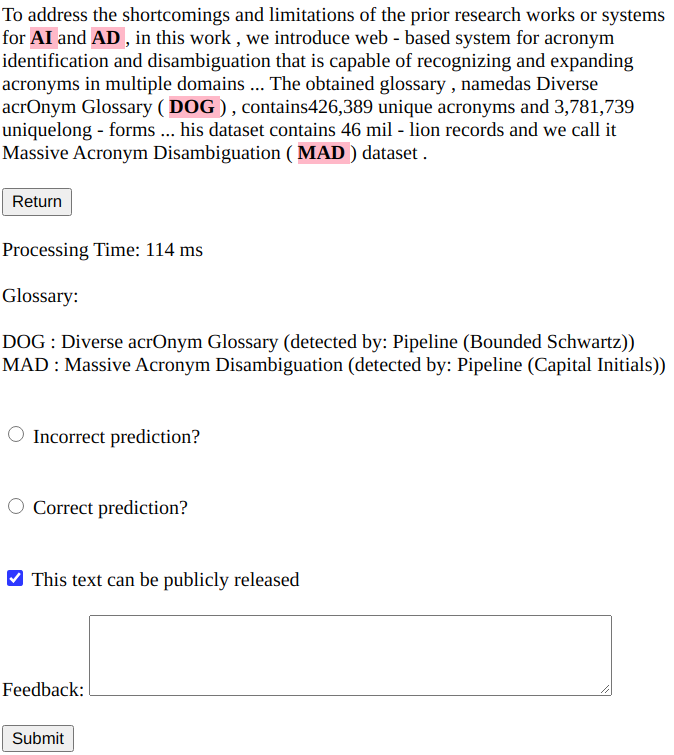
In the web-based system, the user could enter the text and the system recognizes both acronyms without any definition in text and also acronyms that are locally defined with their identified long-forms. Users could also click on each detected acronym to see its definition in a pop-up window. Also, a glossary of detected acronyms and their long-forms is shown at the bottom of the page. A screenshot of the output of the system is shown in Figure 1. Moreover, Table 1 shows the glossary extracted from the text of this paper using the rule-based component of the system. In section 3 we compare the performance of the proposed rule-based model with the existing state-of-the-art models for AI Veyseh et al. (2020).
2.2 Acronym Expansion
Although the proposed rule-based model is effective to recognize locally defined acronyms, it might not be able to expand acronyms that don’t have any immediate definition in the text itself. To alleviate this issue and expand acronyms even without local definition, two resources are required: 1) A dictionary that provides the list of possible expansion for a given acronym, 2) A model to exploit the context of the given acronym and choose the most likely expansion for a given acronym. For the acronym dictionary, we employ the glossary obtained by exploiting our proposed rule-based AI model on corpora in various domains (i.e., Wikipedia, Arxiv papers, Reddit submissions, Medline abstracts, and PMC OA subset). The obtained glossary, named as Diverse acrOnym Glossary (DOG), contains 426,389 unique acronyms and 3,781,739 unique long-forms. Note that the previously available web-based acronym disambiguation system Ciosici and Assent (2018) employed only Wikipedia corpus, therefore, it covers limited domains and acronyms compared to our system.
| Model | Acronym | Long Form | |||||
| P | R | F1 | P | R | F1 | Macro F1 | |
| NOA | 80.31 | 18.08 | 29.51 | 88.97 | 14.01 | 24.20 | 26.85 |
| ADE | 79.28 | 86.13 | 82.57 | 98.36 | 57.34 | 72.45 | 79.37 |
| UAD | 86.11 | 91.48 | 88.72 | 96.51 | 64.38 | 77.24 | 84.09 |
| BIOADI | 83.11 | 87.21 | 85.11 | 90.43 | 73.79 | 77.49 | 82.35 |
| LNCRF | 84.51 | 90.45 | 87.37 | 95.13 | 69.18 | 80.10 | 83.73 |
| LSTM-CRF | 88.58 | 86.93 | 87.75 | 85.33 | 85.38 | 85.36 | 86.55 |
| MadDog | 89.98 | 87.56 | 88.75 | 96.45 | 79.53 | 87.18 | 88.12 |
In DOG, the average number of long-forms per acronym is 6.9 and 81,372 ambiguous acronyms exist. Due to this ambiguity, a simple dictionary look-up is not sufficient for acronym expansion in the web-based system that uses DOG to expand acronyms with non-local definitions. In order to tackle this problem, we propose to train a supervised model in which the input is the text and the position of the ambiguous acronym in it and the model predicts the correct long-form among all possible candidates. To train this model, we use an automatically labeled dataset obtained by extracting samples from large corpora for each long-form in DOG. This dataset contains 46 million records and we call it the Massive Acronym Disambiguation (MAD) dataset. To split the dataset into train/dev/test splits, we use 80% of samples of each long-form for training, 10% for the development set, and 10% for the test set. It is noteworthy that to facilitate training, before splitting the dataset into train/dev/test splits, we first create chunks of size 100,000 samples in which all samples of an acronym are assigned to the same chunk. Since each acronym appears only in one chuck, we train a separate acronym disambiguation model for each chunk. During inference, we first identify which chuck the ambiguous acronym belongs to, then, we use the corresponding model to predict the expanded form of the acronym.
In this work, we use a deep sequential model to be trained on the MAD dataset for acronym disambiguation. More specifically, given the input text with the ambiguous acronym , we first represent each word using the corresponding GloVe embedding, i.e., . Afterward, the vectors are consumed by a Bi-directional Long Short-Term Memory network (BiLSTM) to encode the sequential order of the words. Next, we take the hidden states of the BiLSTM neurons, i.e., , and compute the text representation by computing the max-pool of the vectors , i.e., . Finally, the concatenation of the text representation, i.e., , and the acronym representation, i.e., , is fed into a 2-layer feed-forward neural network whose final layer dimension is equal to the total number of long-forms in the dataset (i.e., dataset chunks explained above).

In the proposed system, the long-form of acronyms predicted by the acronym disambiguation model is presented in the glossary at the end of the page (See Figure 1). Moreover, by clicking on the acronym word in text, a pop-up window shows the model’s prediction and also the sorted list of other candidate long-forms for the selected acronym. An example is shown in Figure 2. In the provided example, the system correctly predicts Gross Domestic Production as the long-form of the ambiguous acronym GDP. We name the proposed acronym identification and disambiguation system as MAdDog.
3 Evaluation
This section provides more insight into the performance of the proposed acronym identification and disambiguation models. To evaluate the performance of the models in comparison with other state-of-the-art AI and AD models, we report the performance of the proposed models on SciAI and SciAD benchmark datasets Veyseh et al. (2020). We also compare the performance of the proposed model with the baselines provided in the recent work Veyseh et al. (2020). More specifically, on SciAI, we compare our model with rule-based models NOA Charbonnier and Wartena (2018), ADE Li et al. (2018) and UAD Ciosici et al. (2019); and also the feature-based models BIOADI Kuo et al. (2009) and LNCRF Liu et al. (2017); and finally the SOTA deep model LSTM-CRF Veyseh et al. (2020). For evaluation metrics, following prior work, we report precision, recall, and F1 score for the acronym and long-form prediction and also their macro-averaged F1 score. The results are shown in Table 2. This table shows that our model outperforms both rule-based and more advanced feature-based or deep learning models. More interestingly, while the proposed model has comparable precision with the existing rule-based models, it enjoys higher recall.
| Model | P | R | F1 |
|---|---|---|---|
| MF | 89.03 | 42.2 | 57.26 |
| ADE | 86.74 | 43.25 | 57.72 |
| NOA | 78.14 | 35.06 | 48.40 |
| UAD | 89.01 | 70.08 | 78.37 |
| BEM | 86.75 | 35.94 | 50.82 |
| DECBAE | 88.67 | 74.32 | 80.86 |
| GAD | 89.27 | 76.66 | 81.90 |
| MadDog | 92.27 | 85.01 | 88.49 |
To assess the performance of the proposed acronym disambiguation model, we evaluate its performance on the benchmark dataset SciAD Veyseh et al. (2020) and compare it with the existing state-of-the-art models. Specifically, we compare the model with non-deep learning models including most frequent (MF) meaning Veyseh et al. (2020), feature-based model (i.e., ADE Li et al. (2018)), and deep learning models including NOA Charbonnier and Wartena (2018), UAD Ciosici et al. (2019), BEM Zettlemoyer and Blevins (2020), DECBAE Jin et al. (2019) and GAD Veyseh et al. (2020). The results are shown in Table 3. This table demonstrates the effectiveness of the proposed model compared with the baselines. Our hypothesis for the higher performance of the proposed model is the massive number of training examples for all acronyms which results in low generalization error.
4 Related Work
Acronym identification (AI) and acronym disambiguation (AD) are two well-known tasks with several prior works in the past two decades. For AI, both rule-based models Park and Byrd (2001); Wren and Garner (2002); Schwartz and Hearst (2002); Adar (2004); Nadeau and Turney (2005); Ao and Takagi (2005); Kirchhoff and Turner (2016) and supervised feature-based or deep learning models Kuo et al. (2009); Liu et al. (2017); Veyseh et al. (2020); Pouran Ben Veyseh et al. (2021) are utilized. Due to the higher accuracy of rule-based models, they are dominantly used in the majority of the related works, especially to automatically create acronym dictionary Ciosici et al. (2019); Li et al. (2018); Charbonnier and Wartena (2018). However, the existing works prepare a small-size dictionary in a specific domain. In contrast, in this work, we first improve the existing rules for acronym identification, then, we use a diverse acronym glossary in our system. For acronym disambiguation, prior works employ either feature-based models Wang et al. (2016); Li et al. (2018) or deep learning methods Wu et al. (2015); Antunes and Matos (2017); Charbonnier and Wartena (2018); Ciosici et al. (2019); Pouran Ben Veyseh et al. (2021). In this work, we also employ a sequential deep learning model for AD. However, unlike prior work that proposes an acronym disambiguation model for a specific domain and limited acronyms, our proposed model covers more acronyms and it is able to expand an acronym in various domains.
Another common limitation of the existing research-based models for AI and AD is that they do not provide any publicly available system that could be quickly incorporated into a text-processing application. Although there is some software for acronym identification such as expanding Biomedical Abbreviations using Dynamic Regular Expressions (BADREX) Gooch (2011) or Abbreviation Expander (ABBREX) ABBREX (2018), unfortunately, they are incapable of acronym disambiguation. To our knowledge, the most similar work to ours is proposed by Ciosici and Assent (2018). Specifically, similar to our work, this web-based system is able to identify and expand acronym in text. A rule-based model is employed for AI and this model is also used to create a dictionary of acronyms. For AD, unlike our work that trains a deep model, they use word embedding similarity to predict the most likely expansion. However, there are some limitations to this previous system. Firstly, it is restricted to the general domain (i.e., Wikipedia) and it covers a limited number of acronyms. Second, it does not provide any analysis and evaluations of the performance of the proposed model. Lastly, it is not publicly available anymore. The proposed MadDog system could be useful for many downstream applications including definition extraction Pouran Ben Veyseh et al. (2020a); Spala et al. (2020, 2019), information extraction Pouran Ben Veyseh et al. (2019, 2020b, 2020c) or question answering Perez et al. (2020)
5 System Deployment
MadDog is purely written in Python 3 and could be run as a FLASK Grinberg (2018) server. For text toknization, it employs SpaCy 2 Honnibal and Montani (2017). Also, the trained acronym expansion model requires PyTorch 1.7 and 64 GB of disk space. Note that all acronyms with their long-forms are encoded in the trained model so they can perform both the dictionary look-up operation and the disambiguation task. Moreover, the trained models could be loaded both on GPU and CPU.
6 Conclusion
In this work, we propose a new web-based system for acronym identification and disambiguation. For AI, we employ a refined set of rules which is shown to be more effective than the previous rule-based and deep learning models. Moreover, using a massive acronym disambiguation dataset with more than 46 million records in various domains, we train a supervised model for acronym disambiguation. The experiments on the existing benchmark datasets reveal the efficacy of the proposed AD model.
References
- ABBREX (2018) ABBREX2018. Abbrex. 2018. abbrex - the abbreviation expander. In BMC bioinformatics.
- Adar (2004) Eytan Adar. 2004. Sarad: A simple and robust abbreviation dictionary. In Bioinformatics.
- Antunes and Matos (2017) Rui Antunes and Sérgio Matos. 2017. Biomedical word sense disambiguation with word embeddings. In International Conference on Practical Applications of Computational Biology & Bioinformatics.
- Ao and Takagi (2005) Hiroko Ao and Toshihisa Takagi. 2005. Alice: an algorithm to extract abbreviations from medline. In Journal of the American Medical Informatics Association.
- Charbonnier and Wartena (2018) Jean Charbonnier and Christian Wartena. 2018. Using word embeddings for unsupervised acronym disambiguation. In Proceedings of the 27th International Conference on Computational Linguistics.
- Ciosici and Assent (2018) Manuel R. Ciosici and Ira Assent. 2018. Abbreviation expander - a web-based system for easy reading of technical documents. In Proceedings of the 27th International Conference on Computational Linguistics: System Demonstrations.
- Ciosici et al. (2019) Manuel R Ciosici, Tobias Sommer, and Ira Assent. 2019. Unsupervised abbreviation disambiguation. In arXiv preprint arXiv:1904.00929.
- Gooch (2011) Phil Gooch. 2011. Badrex: In situ expansion and coreference of biomedical abbreviations using dynamic regular expressions.
- Grinberg (2018) Miguel Grinberg. 2018. Flask web development: developing web applications with python. ” O’Reilly Media, Inc.”.
- Honnibal and Montani (2017) Matthew Honnibal and Ines Montani. 2017. spaCy 2: Natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsing. To appear.
- Jin et al. (2019) Qiao Jin, Jinling Liu, and Xinghua Lu. 2019. Deep contextualized biomedical abbreviation expansion. In arXiv preprint arXiv:1906.03360.
- Kirchhoff and Turner (2016) Katrin Kirchhoff and Anne M Turner. 2016. Unsupervised resolution of acronyms and abbreviations in nursing notes using document-level context models. In Proceedings of the Seventh International Workshop on Health Text Mining and Information Analysis.
- Kuo et al. (2009) Cheng-Ju Kuo, Maurice HT Ling, Kuan-Ting Lin, and Chun-Nan Hsu. 2009. Bioadi: a machine learning approach to identifying abbreviations and definitions in biological literature. In BMC bioinformatics.
- Li et al. (2018) Yang Li, Bo Zhao, Ariel Fuxman, and Fangbo Tao. 2018. Guess me if you can: Acronym disambiguation for enterprises. In ACL.
- Liu et al. (2017) Jie Liu, Caihua Liu, and Yalou Huang. 2017. Multi-granularity sequence labeling model for acronym expansion identification. In Information Sciences.
- Nadeau and Turney (2005) David Nadeau and Peter D Turney. 2005. A supervised learning approach to acronym identification. In Conference of the Canadian Society for Computational Studies of Intelligence.
- Park and Byrd (2001) Youngja Park and Roy J Byrd. 2001. Hybrid text mining for finding abbreviations and their definitions. In Proceedings of the 2001 conference on empirical methods in natural language processing.
- Perez et al. (2020) Ethan Perez, Patrick Lewis, Wen-tau Yih, Kyunghyun Cho, and Douwe Kiela. 2020. Unsupervised question decomposition for question answering. In EMNLP.
- Pouran Ben Veyseh et al. (2020a) Amir Pouran Ben Veyseh, Franck Dernoncourt, Dejing Dou, and Thien Huu Nguyen. 2020a. A joint model for definition extraction with syntactic connection and semantic consistency. In AAAI.
- Pouran Ben Veyseh et al. (2020b) Amir Pouran Ben Veyseh, Franck Dernoncourt, Dejing Dou, and Thien Huu Nguyen. 2020b. Exploiting the syntax-model consistency for neural relation extraction. In ACL.
- Pouran Ben Veyseh et al. (2021) Amir Pouran Ben Veyseh, Franck Dernoncourt, Thien Huu Nguyen, Walter Chang, and Leo Anthony Celi. 2021. Acronym identification and disambiguation shared tasks for scientific document understanding. In Proceedings of the 1st workshop on Scientific Document Understanding at AAAI-21.
- Pouran Ben Veyseh et al. (2020c) Amir Pouran Ben Veyseh, Franck Dernoncourt, My Thai, Dejing Dou, and Thien Nguyen. 2020c. Multi-view consistency for relation extraction via mutual information and structure prediction. In AAAI.
- Pouran Ben Veyseh et al. (2019) Amir Pouran Ben Veyseh, Thien Nguyen, and Dejing Dou. 2019. Improving cross-domain performance for relation extraction via dependency prediction and information flow control. In IJCAI.
- Schwartz and Hearst (2002) Ariel S Schwartz and Marti A Hearst. 2002. A simple algorithm for identifying abbreviation definitions in biomedical text. In Biocomputing 2003.
- Spala et al. (2020) Sasha Spala, Nicholas Miller, Franck Dernoncourt, and Carl Dockhorn. 2020. SemEval-2020 task 6: Definition extraction from free text with the DEFT corpus. In Proceedings of the Fourteenth Workshop on Semantic Evaluation.
- Spala et al. (2019) Sasha Spala, Nicholas A. Miller, Yiming Yang, Franck Dernoncourt, and Carl Dockhorn. 2019. DEFT: A corpus for definition extraction in free- and semi-structured text. In Proceedings of the 13th Linguistic Annotation Workshop.
- Veyseh et al. (2020) Amir Pouran Ben Veyseh, Franck Dernoncourt, Quan Hung Tran, and Thien Huu Nguyen. 2020. What Does This Acronym Mean? Introducing a New Dataset for Acronym Identification and Disambiguation. In Proceedings of COLING.
- Wang et al. (2016) Yue Wang, Kai Zheng, Hua Xu, and Qiaozhu Mei. 2016. Clinical word sense disambiguation with interactive search and classification. In AMIA Annual Symposium Proceedings.
- Wen et al. (2020) Zhi Wen, Xing Han Lu, and Siva Reddy. 2020. MeDAL: Medical abbreviation disambiguation dataset for natural language understanding pretraining. In Proceedings of the 3rd Clinical Natural Language Processing Workshop.
- Wren and Garner (2002) Jonathan D Wren and Harold R Garner. 2002. Heuristics for identification of acronym-definition patterns within text: towards an automated construction of comprehensive acronym-definition dictionaries. In Methods of information in medicine.
- Wu et al. (2015) Yonghui Wu, Jun Xu, Yaoyun Zhang, and Hua Xu. 2015. Clinical abbreviation disambiguation using neural word embeddings. In Proceedings of BioNLP 15.
- Zettlemoyer and Blevins (2020) Luke Zettlemoyer and Terra Blevins. 2020. Moving down the long tail of word sense disambiguation with gloss informed bi-encoders. In ACL.