Literature Retrieval for Precision Medicine
with Neural Matching and Faceted Summarization
Abstract
Information retrieval (IR) for precision medicine (PM) often involves looking for multiple pieces of evidence that characterize a patient case. This typically includes at least the name of a condition and a genetic variation that applies to the patient. Other factors such as demographic attributes, comorbidities, and social determinants may also be pertinent. As such, the retrieval problem is often formulated as ad hoc search but with multiple facets (e.g., disease, mutation) that may need to be incorporated. In this paper, we present a document reranking approach that combines neural query-document matching and text summarization toward such retrieval scenarios. Our architecture builds on the basic BERT model with three specific components for reranking: (a). document-query matching (b). keyword extraction and (c). facet-conditioned abstractive summarization. The outcomes of (b) and (c) are used to essentially transform a candidate document into a concise summary that can be compared with the query at hand to compute a relevance score. Component (a) directly generates a matching score of a candidate document for a query. The full architecture benefits from the complementary potential of document-query matching and the novel document transformation approach based on summarization along PM facets. Evaluations using NIST’s TREC-PM track datasets (2017–2019) show that our model achieves state-of-the-art performance. To foster reproducibility, our code is made available here: https://github.com/bionlproc/text-summ-for-doc-retrieval.
1 Introduction
The U.S. NIH’s precision medicine (PM) initiative Collins and Varmus (2015) calls for designing treatment and preventative interventions considering genetic, clinical, social, behavioral, and environmental exposure variability among patients. The initiative rests on the widely understood finding that considering individual variability is critical in tailoring healthcare interventions to achieve substantial progress in reducing disease burden worldwide. Cancer was chosen as its near term focus with the eventual aim of expanding to other conditions. As the biomedical research enterprise strives to fulfill the initiative’s goals, computing needs are also on the rise in drug discovery, predictive modeling for disease onset and progression, and in building NLP tools to curate information from the evidence base being generated.
1.1 TREC Precision Medicine Series
| Facet | Input |
|---|---|
| Disease | Melanoma |
| Genetic variation | BRAF (E586K) |
| Demographics | 64-year-old female |
| Disease | Gastric cancer |
| Genetic variation | ERBB2 amplification |
| Demographics | 64-year-old male |
In a dovetailing move, the U.S. NIST’s TREC (Text REtrieval Conference) has been running a PM track since 2017 with a focus on cancer Roberts et al. (2020). The goal of the TREC-PM task is to identify the most relevant biomedical articles and clinical trials for an input patient case. Each case is composed of (1) a disease name, (2) a gene name and genetic variation type, and (3) demographic information (sex and age). Table 1 shows two example cases from the 2019 track. So the search is ad hoc in the sense that we have a free text input in each facet but the facets themselves highlight the PM related attributes that ought to characterize the retrieved documents. We believe this style of faceted retrieval is going to be more common across medical IR tasks for many conditions as the PM initiative continues its mission.
1.2 Vocabulary Mismatch and Neural IR
The vocabulary mismatch problem is a prominent issue in medical IR given the large variation in the expression of medical concepts and events. For example, in the query “What is a potential side effect for Tymlos?” the drug is referred by its brand name. Relevant scientific literature may contain the generic name Abaloparatide more frequently. Traditional document search engines have clear limitations on resolving mismatch issues. The IR community has extensively explored methods to address the vocabulary mismatch problem, including query expansion based on relevance feedback, query term re-weighting, or query reconstruction by optimizing the query syntax.
Several recent studies highlight exploiting neural network models for query refinement in document retrieval (DR) settings. Nogueira and Cho (2017) address this issue by generating a transformed query from the initial query using a neural model. They use reinforcement learning (RL) to train it where an agent (i.e., reformulator) learns to reformulate the initial query to maximize the expected return (i.e., retrieval performance) through actions (i.e., generating a new query from the output probability distribution). In a different approach, Narayan et al. (2018) use RL for sentence ranking for extractive summarization.
1.3 Our Contributions
In this paper, building on the BERT architecture Devlin et al. (2019), we focus on a different hybrid document scoring and reranking setup involving three components: (a). a document relevance classification model, which predicts (and inherently scores) whether a document is relevant to the given query (using a BERT multi-sentence setup); (b). a keyword extraction model which spots tokens in a document that are likely to be seen in PM related queries; and (c). an abstractive document summarization model that generates a pseudo-query given the document context and a facet type (e.g., genetic variation) via the BERT encoder-decoder setup. The keywords (from (b)) and the pseudo-query (from (c)) are together compared with the original query to generate a score. The scores from all the components are combined to rerank top (set to 500) documents returned with a basic Okapi BM25 retriever from a Solr index Grainger and Potter (2014) of the corpora.
Our main innovation is in pivoting from the focus on queries by previous methods to emphasis on transforming candidate documents into pseudo-queries via summarization. Additionally, while generating the pseudo-query, we also let the decoder output concept codes from biomedical terminologies that capture disease and gene names. We do this by embedding both words and concepts in a common semantic space before letting the decoder generate summaries that include concepts. Our overall architecture was evaluated using the TREC-PM datasets (2017–2019) with the 2019 dataset used as the test set. The results show an absolute improvement in P@10 compared to prior best approaches while obtaining a small gain in R-Prec. Qualitative analyses also highlight how the summarization is able to focus on document segments that are highly relevant to patient cases.
2 Background
The basic reranking architecture we begin with is the Bidirectional Encoder Representations from Transformers (BERT) Devlin et al. (2019) model. BERT is trained on a masked language modeling objective on a large text corpus such as Wikipedia and BooksCorpus. As a sequence modeling method, it has achieved state-of-the-art results in a wide range of natural language understanding (NLU) tasks, including machine translation Conneau and Lample (2019) and text summarization Liu and Lapata (2019). With an additional layer on top of a pretrained BERT model, we can fine-tune models for specific NLU tasks. In our study, we utilize this framework in all three components identified in Section 1.3 by starting with a bert-base-uncased pretrained HuggingFace model Wolf et al. (2019).
2.1 Text Summarization
We plan to leverage both extractive and abstractive candidate document summarization in our framework. In terms of learning methodology, we view extractive summarization as a sentence (or token) classification problem. Previously proposed models include the RNN-based sequence model Nallapati et al. (2017), the attention-based neural encoder-decoder model Cheng and Lapata (2016), and the sequence model with a global learning objective (e.g., ROUGE) for ranking sentences optimized via RL Narayan et al. (2018); Paulus et al. (2018). More recently, graph convolutional neural networks (GCNs) have also been adapted to allow the incorporation of global information in text summarization tasks Sun et al. (2019); Prasad and Kan (2019). Abstractive summarization is typically cast as a sequence-to-sequence learning problem. The encoder of the framework reads a document and yields a sequence of continuous representations, and the decoder generates the target summary token-by-token Rush et al. (2015); Nallapati et al. (2016). Both approaches have their own merits in generating comprehensive and novel summaries; hence most systems leverage these two different models in one framework See et al. (2017); Liu and Lapata (2019). We use the extractive component to identify tokens in a candidate document that may be relevant from a PM perspective and use the abstractive component to identify potential terms that may not necessarily be in the document but nevertheless characterize it for PM purposes.
2.2 Word and Entity Embeddings
Most of the neural text summarization models, as described in the previous section, adopt the encoder-decoder framework that is popular in machine translation. As such the vocabulary on the decoding side does not have to be the same as that on the encoding side. We exploit this to design a summarization trick for PM where the decoder outputs both regular English tokens and also entity codes from a standardized biomedical terminology that captures semantic concepts discussed in the document. This can be trained easily by converting the textual queries in the training examples to their corresponding entity codes. This trick is to enhance our ability to handle vocabulary mismatch in a different way (besides the abstractive framing). We created BioMedical Entity Tagged (BMET) embeddings111https://github.com/romanegloo/BMET-embeddings for this purpose. BMET embeddings are trained on biomedical literature abstracts that were annotated with entity codes in the Medical Subject Headings (MeSH) terminology222https://www.nlm.nih.gov/mesh/meshhome.html; codes are appended to the associated textual spans in the training examples. So regular tokens and the entity codes are thus embedded in the same semantic space via pretraining with the fastText architecture Bojanowski et al. (2017). Besides regular English tokens, the vocabulary of BMET thus includes 29,351 MeSH codes and a subset of supplementary concepts. In the dictionary, MeSH codes are differentiated from the regular words by a unique prefix; for example, mesh_d000123 for MeSH code D000123. With this, our summarization model can now translate a sequence of regular text tokens into a sequence of biomedical entity codes or vice versa. That is, we use MeSH as a new “semantic” facet besides those already provided by TREC-PM organizers. The expected output for the MeSH facet is the set of codes that capture entities in the disease and gene variation facets.
3 Models and Reranking
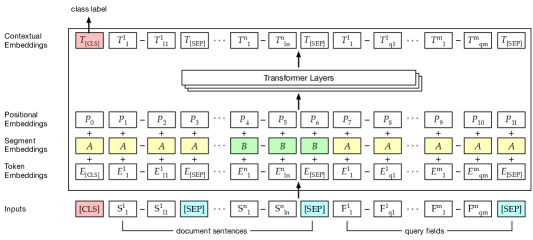
In this effort, toward document reranking, we aim to measure the relevance match between a document and a faceted PM query. Each training instance is a 3-tuple (, , ) where is a query, is a candidate document, and is a Boolean human adjudicated outcome: whether is relevant to . As mentioned in Section 1.3, first, we fine-tune BERT for a query-document relevance matching task modeled as a classification goal to predict (REL). Next, we fine-tune BERT for token-level relevance classification, different from REL, where a token in is deemed relevant during training if it occurs as part of . We name this model EXT for keyword extraction. Lastly, we train a BERT model in the seq2seq setting where the encoder is initialized with a pretrained EXT model. The encoder reads in , and the decoder attends to the contextualized representations of to generate a facet-specific pseudo-query sentence , which is then compared with the original query . We conceptualize this process as text summarization from a document to query sentences333We note queries here are not grammatically well-formed sentences but are essentially sequences generated by the summarization model. and refer to it as ABS. All three models are used together to rerank a candidate at test time for a specific input query.
3.1 Document Relevance Matching (REL)
Neural text matching has been recently carried out through siamese style networks Mueller and Thyagarajan (2016), which also have been adapted to biomedicine Noh and Kavuluru (2018). Our approach adapts the BERT architecture for the matching task in the multi-sentence setting as shown in Figure 1. We use BERT’s tokenizer on its textual inputs, and the tokens are mapped to token embeddings. REL takes the concatenated sequence of a document and faceted query sentences. The functional symbols defined in the BERT tokenizer (e.g., [CLS]) are added to the input sequence. Each input sequence starts with a [CLS] token. Each sentence of the document ends with the [SEP] token with the last segment of the input sequence being the set of faceted query sentences, which end with another [SEP] token. In the encoding process, the first [CLS] token collects features for determining document relevance to the query. BERT uses segment embeddings to distinguish two sentences. We, however, use the them to distinguish multiple sentences within a document. For each sentence, we assign a segment embedding either or alternatively. The positional embeddings encode the sequential nature of the inputs. The token embeddings along with the segment and positional embeddings pass through the transformer layers. Finally, we use the output logit from the [CLS] token () as the matching score for the input document and query. We note that we don’t demarcate any boundaries within different facets of the query.
3.2 Keyword Extraction (EXT)
EXT model has an additional token classification layer on top of the pretrained BERT. The output of a token is the logit that indicates the log of odds of the token’s occurrence in the query. With TREC-PM datasets, we expect to see the logits fire for words related to different facets with an optimized EXT at test time. Unlike the REL model, the input to EXT is a sequence of words in a document without any [SEP] delimiters. However, the model still learns the boundaries of the sentence via segment inputs. This component essentially generates a brief extractive summary of a candidate document. Furthermore, contextualized embeddings from EXT are used in the decoder of ABS to generate faceted abstractive document summaries .
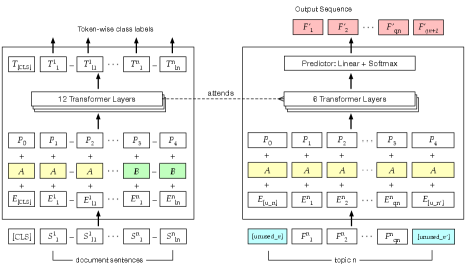
3.3 Abstractive Document Summarization (ABS)
ABS employs a standard seq2seq attention model, similar to that by Nallapati et al. (2016), as shown in Figure 2. We initialize the parameters of the encoder with a pretrained EXT model. The decoder is a 6-layer transformer in which the self-attention layers attend to only the earlier positions in the output sequence as is typical in auto-regressive language models. In each training phase step, the decoder takes each previous token from the reference query sentence; in the generation process, the decoder uses the token predicted one step earlier.
| Facets | (bos)/(eos) |
|---|---|
| Disease name | [unused_0]/[unused_100] |
| Genetic variations | [unused_1]/[unused_101] |
| Demographic info. | [unused_2]/[unused_102] |
| MeSH terms | [unused_3]/[unused_103] |
| Document keywords | [unused_4]/[unused_104] |
We differentiate facets by the special pairs of tokens assigned to each topic. In a typical generation process, special tokens such as [bos] (begin) and [eos] (end) are used to indicate sequence boundaries. In this model, we use some special tokens in the BERT vocabulary with prefix ‘unused_’. Specifically, [unused_] and [unused_] are used as bos and eos tokens respectively for different facets. These facet signals are the latent variables for which ABS is optimized. Through them, ABS learns not only the thematic aspects of the queries but also the meta attributes such as length. The special tokens for facets are listed in Table 2 (the last row indicates a new auxiliary facet we introduce in Section 4.1).
Each faceted query is enclosed by its assigned bos/eos pair, and the decoder of ABS learns , where is the facet signal. As in the encoder and the original transformer architecture Vaswani et al. (2017), we add the sinusoidal positional embedding and the segment vector (or ) to the token embedding . Note that the dimension of the token embeddings used in the encoder (BERT embeddings) is different from that of the decoder (our custom BMET embeddings), which causes a discrepancy in computing context-attentions of the target text across the source document. Hence, we add an additional linear layer to project the constructed decoder embeddings ( in the right hand portion of Figure 2) into the same space of embeddings of the encoder. These projected embeddings are fed to the decoder’s transformer layers. Each transformer layer applies multi-head attention for computing the self- and context-attentions. The attention function reads the input masks to preclude attending to future tokens of the input and any padded tokens (i.e., [PAD]) of the source text. Both attention functions apply a residual connection He et al. (2016). Lastly, each transformer layer ends with a position-wise feedforward network. Final scores for each token are computed from the linear layer on top of the transformer layers. In training, these scores are consumed by a cross-entropy loss function. In generation process, the softmax function is applied over the vocabulary yielding a probability distribution for sampling the next token.
Finally to generate the pseudo-query, we use beam search to find the most probable sentence among predicted candidates. The scores are penalized by two measures proposed by Wu et al. (2016, Equation 14): (1). The length penalty , where is the current target length and is the length normalization coefficient. (2). The coverage penalty
where is the attention score of the -th target word on the -th source word , is the source length, and is the coverage normalization coefficient. Intuitively, these functions avoid favoring shorter predictions and yielding duplicate terms. We tune the parameters of the penalty functions (), with grid-search on the validation set for TREC-PM.
3.4 Reranking with REL, EXT, and ABS
The main purpose of the models designed in the previous subsections is to come up with a combined measure for reranking. For a query , let , be the set of top (set to 500) candidate documents returned by the Solr BM25 eDisMax query. It is straightforward to impose an order on through REL via the output probability estimates of relevance. Given , for each we generate the pseudo-query (summary) by concatenating all distinct words in the generated pseudo-query sentences by ABS along with the words selected by EXT. Repeating words and special tokens are removed. Although faceted summaries are generated through ABS, in the end is essentially the set of all unique terms from ABS and EXT. Each is now scored by comparing and via two similarity metrics: The ROUGE-1 recall score, Lin (2004), and a cosine similarity based score computed as
where denote vector representations from BMET embeddings (Section 2.2).
Overall, we compute four different scores (and hence rankings) of a document: (1) the retrieval score returned by Solr, (2) the document relevance score by REL, (3) pseudo-query based ROUGE score, and (4) pseudo-query similarity score . In the end we merge the rankings with reciprocal rank fusion Cormack et al. (2009) to obtain the final ranked list of documents. The results are compared against the state-of-the-art models from the 2019 TREC-PM task.
4 Experimental Setup
4.1 Data
Across 2017–2019 TREC-PM tasks, we have a total of 120 patient cases and 63,387 qrels (document relevance judgments) as shown in Table 3.
| Year | Queries | Documents (rel. / irrel.) |
|---|---|---|
| 2017 | 30 | 3,875 / 18,767 |
| 2018 | 50 | 5,588 / 16,841 |
| 2019 | 40 | 5,544 / 12,772 |
We create two new auxiliary facets, MeSH terms and Keywords, derived from any training query and document pair. We already covered the MeSH facet in Section 2.2. Keywords are those assigned by authors to a biomedical article to capture its themes and are downloadable from NIH’s NCBI website. If no keywords were assigned to an article, then we use the set of preferred names of MeSH terms (assigned to the articles by trained NIH coders) for that example. The following list shows associated facets for a sample training instance:
-
•
Disease: prostate cancer
-
•
Genetic variations: ATM deletion
-
•
Demographics: 50-year-old male
-
•
MeSH terms: D011471, D064007
-
•
Keywords: Aged, Ataxia Telangiectasia mutated Proteins, Prostate Neoplasms/genetics
Each model consumes data differently, as shown in Table 4. REL takes a document along with the given query as the source input and predicts document-level relevance. We consider a document with the human judgment score either 1 (partially relevant) or 2 (totally relevant) as relevant for this study. Note that we do not include MeSH terms in the query sentences for REL. EXT reads in a document as the source input and predicts token-level relevances. During training, a relevant token is one that occurs in the given patient case. A pseudo-query is the output for ABS taking in a document and a facet type.
| Model | Source | Target |
|---|---|---|
| REL | docquery_sentences | doc relevance |
| EXT | doc | token relevances |
| ABS | docfacet_signal | a pseudo-query |
4.2 Implementation Details
For all three models, we begin with the pretrained bert-base-uncased HuggingFace model Wolf et al. (2019) to encode source texts. We use BERT’s WordPiece Schuster and Nakajima (2012) tokenizer for the source documents.
REL and EXT are trained for 30,000 steps with batch size of 12. The maximum number of tokens for source texts is limited to 384. As the loss function of these two models, we use weighted binary cross entropy. That is, given high imbalance with many more irrelevant instances than positive ones, we put different weights on the classes in computing the loss according to the target distributions (proportions of negative examples are 87% for REL and 93% for EXT). The loss is
where for REL and for EXT. Adam optimizer with parameters and , starting learning rate , and fixed weight decay of 0.0 was used. The learning rate is reduced when a metric has stopped improving by using the ReduceLROnPlateau scheduler in PyTorch.
For the decoder of ABS, multi-head attention module from OpenNMT Klein et al. (2017) was used. To tokenize target texts, we use the NLTK word tokenizer (https://www.nltk.org/api/nltk.tokenize.html) unlike the one used in the encoder; this is because we use customized word embeddings, the BMET embeddings (Section 2.2), trained with a domain-specific corpus and vocabulary. The vocabulary size is 120,000 which includes the 29,351 MeSH codes. We use six transformer layers in the decoder. Model dimension is 768 and the feed-forward layer size is 2048. We use different initial learning rates for the encoder and decoder, since the encoder is initialized with a pretrained EXT model: (encoder) and (decoder). Negative log-likelihood is the loss function for ABS on the ground-truth faceted query sentences. For beam search in ABS, beam_size is set to 4. At test time, we select top two best predictions and merge them into one query sentence. The max length of target sentence is limited to 50 and a sequence is incrementally generated until ABS outputs the corresponding eos token for each facet. All parameter choices were made based on best practices from prior efforts and experiments to optimize P@10 on validation subsets.
5 Evaluations and Results
We conducted both quantitative and qualitative evaluations with example outcomes. The final evaluation was done on the 2019 TREC-PM dataset while all hyperparameter tuning was done using a training and validation dataset split of a shuffled combined set of instances from 2017 and 2018 tracks (20% validation and the rest for training).
5.1 Quantitative Evaluations
We first discuss the performances of the constituent REL and EXT models that were evaluated using train and validation splits from 2017–2018 years. Table 5 shows their performance where REL can recover of the relevant documents and EXT can identify of the tokens that occur in patient case information, both at precisions over 90%. We find that learning a model for identifying document/token-level relevance is relatively straightforward even with the imbalance.
| REL | EXT | |||||
|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | |
| Train | 0.9814 | 0.9384 | 0.9594 | 0.9624 | 0.8877 | 0.9236 |
| Valid | 0.9266 | 0.9147 | 0.9206 | 0.9413 | 0.8732 | 0.9060 |
Next we discuss the main results comparing against the top two teams (rows 1–2) in the 2019 track in Table 6. Before we proceed, we want to highlight one crucial evaluation consideration that applies to any TREC track. TREC evaluates systems in the Cranfield paradigm where pooled top documents from all participating teams are judged for relevance by human experts. Because we did not participate in the original TREC-PM 2019 task, our retrieved results are not part of the judged documents. Hence, we may be at a slight disadvantage when comparing our results with those of teams that participated in 2019 TREC-PM. Nevertheless, we believe that at least the top few most relevant documents are typically commonly retrieved by all models. Hence we compare with both P@10 and R-Prec (P@all-relevant-doc-count) measures.
| Model | R-Prec | P@10 |
|---|---|---|
| julie-mug Faessler et al. (2020) | 0.3572 | 0.6525 |
| BITEM_PM Caucheteur et al. (2020) | 0.3166 | 0.6275 |
| Baseline: Solr eDisMax | 0.2307 | 0.5200 |
| Baseline + Solr MLT | 0.1773 | 0.2625 |
| Baseline + REL | 0.3912 | 0.6750 |
| Baseline + ABS | 0.2700 | 0.5625 |
| Baseline + REL+ABS | 0.3627 | 0.6985 |
Our baseline Solr query results are shown in row 3 with subsequent rows showing results from additional components. Solr eDisMax is a document ranking function which is based on the BM25 Jones et al. (2000) probabilistic model. We also evaluate eDisMax with Solr MLT (MoreLikeThis), in which a new query is generated by adding a few “interesting” terms (top TF/IDF terms) from the retrieved documents of the initial eDisMax query. This traditional relevance feedback method (row 4) method has decreased the performance from the baseline and hence has not been used in our reranking methods.
All our models (rows 5–7) present stable baseline scores in P@10 and the combined method (+REL+ABS) tops the list with a 4% improvement over the prior best model Faessler et al. (2020). Baseline with REL does the best in terms of R-Prec. Both prior top teams rely heavily on query expansion through external knowledge bases to add synonyms, hypernyms, and hyponyms of terms found in the original query.
5.2 Qualitative Analysis
| Document | Facet signal | Summary |
|---|---|---|
| Title: Association between BRAF v600e mutation and the clinicopathological features of solitary papillary thyroid microcarcinoma. (PMID: 28454296) | [unused_0] | papillary intrahepatic cholangiocarcinoma |
| [unused_1] | braf v600e | |
| [unused_3] | D018281 C535533 | |
| [unused_4] | papillary thyroid braf clinicopathological v600e | |
| Title: Identification of differential and functionally active miRNAs in both anaplastic lymphoma kinase (ALK)+ and ALK- anaplastic large-cell lymphoma. (PMID: 20805506) | [unused_0] | lymphoma |
| [unused_1] | anaplastic lymphoma alk cell bradykinin | |
| [unused_3] | D002471 D017728 D000077548 | |
| [unused_4] | lymphoma alk receptor tyrosine kinase |
Table 7 presents sample pseudo-queries generated by ABS. The summaries of the first document show some novel words, intrahepatic and cholangiocarcinoma, that do not occur in the given document (we only show title for conciseness, but the abstract also does not contain those words). The model may have learned the close relationship between cholangiocarcinoma and BRAF v600e, the latter being part of the genetic facet of the actual query for which PMID: 28454296 turns out to be relevant. Also embedding proximity between intrahepatic and cholangiocarcinoma may have introduced both into the pseudo query, although they are not central to this document’s theme. Still, this maybe important in retrieving documents that have an indirect (yet relevant) link to the query through the pseudo-query terms. This could be why, although ABS underperforms REL, it still complements it when combined (Table 6). The table also shows that ABS can generate concepts in a domain-specific terminology. For example, the second document yields following MeSH entity codes, which are strongly related to the topics of the document: D002471 (Cell Transformation, Neoplastic), D017728 (Lymphoma, Large-Cell, Anaplastic), and D000077548 (Anaplastic Lymphoma Kinase).
For a qualitative exploration of what EXT and different facets of ABS capture, we refer the reader to Appendix A.
5.3 Machine Configuration and Runtime
All training and testing was done on a single Nvidia Titan X GPU in a desktop with 64GB RAM. The corpus to be indexed had 30,429,310 biomedical citations (titles and abstracts of biomedical articles444Due to copyright issues with full-text, TREC-PM is only conducted on abstracts/titles of articles available on PubMed.). We trained the three models for five epochs and the training time per epoch (80,319 query, doc pairs) is 69 mins for REL, 72 mins for EXT, and 303 mins for ABS. Coming to test time, per query, the Solr eDisMax query returns top 500 results in 20 ms. Generating pseudo-queries for 500 candidates via EXT and ABS takes 126 seconds and generating REL scores consumes 16 seconds. So per query, it takes nearly 2.5 mins at test time to return a ranked list of documents. Although this does not facilitate real time retrieval as in commercial search engines, given the complexity of the queries, we believe this is at least near real time offering a convenient way to launch PM queries. Furthermore, this comes at an affordable configuration for many labs and clinics with a smaller carbon footprint.
6 Conclusion
In this paper, we proposed an ensemble document reranking approach for PM queries. It builds on pretrained BERT models to combine strategies from document relevance matching and extractive/abstractive text summarization to arrive at document rankings that are complementary in eventual evaluations. Our experiments also demonstrate that entity embeddings trained on an annotated domain specific corpus can help in document retrieval settings. Both quantitative and qualitative analyses throw light on the strengths of our approach.
One scope for advances lies in improving the summarizer to generate better pseudo-queries so that ABS starts to perform better on its own. At a high level, training data is very hard to generate in large amounts for IR tasks in biomedicine and this holds for the TREC-PM datasets too. To better train ABS, it may be better to adapt other biomedical IR datasets. For example, the TREC clinical decision support (CDS) task that ran from 2014 to 2016 is related to the PM task Roberts et al. (2016). A future goal is to see if we can apply our neural transfer learning Rios and Kavuluru (2019) and domain adaptation Rios et al. (2018) efforts to repurpose the CDS datasets for the PM task.
Another straightforward idea is to reuse generated pseudo-query sentences in the eDisMax query by Solr, as a form of pseudo relevance feedback. The expression in Section 3.4 focuses on an asymmetric formulation that starts with a query term and looks for the best match in the pseudo-query. Considering a more symmetric formulation, where, we also begin with the pseudo-query terms and average both summands may provide a better estimate for reranking. Additionally, a thorough exploration of how external biomedical knowledge bases Wagner et al. (2020) can be incorporated in the neural IR framework for PM is also important Nguyen et al. (2017).
References
- Bojanowski et al. (2017) Piotr Bojanowski, Edouard Grave, Armand Joulin, and Tomas Mikolov. 2017. Enriching word vectors with subword information. Transactions of the Association for Computational Linguistics, 5:135–146.
- Caucheteur et al. (2020) Deborah Caucheteur, Emilie Pasche, Julien Gobeill, Anais Mottaz, Luc Mottin, and Patrick Ruch. 2020. Designing retrieval models to contrast precision-driven ad hoc search vs. recall-driven treatment extraction in precision medicine.
- Cheng and Lapata (2016) Jianpeng Cheng and Mirella Lapata. 2016. Neural summarization by extracting sentences and words. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 484–494.
- Collins and Varmus (2015) Francis S Collins and Harold Varmus. 2015. A new initiative on precision medicine. New England journal of medicine, 372(9):793–795.
- Conneau and Lample (2019) Alexis Conneau and Guillaume Lample. 2019. Cross-lingual language model pretraining. In Advances in Neural Information Processing Systems, pages 7057–7067.
- Cormack et al. (2009) Gordon V Cormack, Charles LA Clarke, and Stefan Buettcher. 2009. Reciprocal rank fusion outperforms condorcet and individual rank learning methods. In Proceedings of the 32nd international ACM SIGIR conference on Research and development in information retrieval, pages 758–759.
- Devlin et al. (2019) Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. 2019. BERT: Pre-training of deep bidirectional transformers for language understanding. In NAACL-HLT, pages 4171–4186.
- Faessler et al. (2020) Erik Faessler, Michel Oleynik, and Udo Hahn. 2020. JULIE lab & Med Uni Graz @ TREC 2019 precision medicine track.
- Grainger and Potter (2014) Trey Grainger and Timothy Potter. 2014. Solr in action. Manning Publications Co.
- He et al. (2016) Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. 2016. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 770–778.
- Jones et al. (2000) K Sparck Jones, Steve Walker, and Stephen E. Robertson. 2000. A probabilistic model of information retrieval: development and comparative experiments: Part 2. Information processing & management, 36(6):809–840.
- Klein et al. (2017) Guillaume Klein, Yoon Kim, Yuntian Deng, Jean Senellart, and Alexander M Rush. 2017. Opennmt: Open-source toolkit for neural machine translation. In Proceedings of ACL 2017, System Demonstrations, pages 67–72.
- Lin (2004) Chin-Yew Lin. 2004. ROUGE: A package for automatic evaluation of summaries. In Text Summarization Branches Out, pages 74–81, Barcelona, Spain. Association for Computational Linguistics.
- Liu and Lapata (2019) Yang Liu and Mirella Lapata. 2019. Text summarization with pretrained encoders. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), pages 3721–3731.
- Mueller and Thyagarajan (2016) Jonas Mueller and Aditya Thyagarajan. 2016. Siamese recurrent architectures for learning sentence similarity. In Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence, pages 2786–2792.
- Nallapati et al. (2017) Ramesh Nallapati, Feifei Zhai, and Bowen Zhou. 2017. Summarunner: A recurrent neural network based sequence model for extractive summarization of documents. In Thirty-First AAAI Conference on Artificial Intelligence.
- Nallapati et al. (2016) Ramesh Nallapati, Bowen Zhou, Cicero dos Santos, Çağlar Gulçehre, and Bing Xiang. 2016. Abstractive text summarization using sequence-to-sequence rnns and beyond. In Proceedings of The 20th SIGNLL Conference on Computational Natural Language Learning, pages 280–290.
- Narayan et al. (2018) Shashi Narayan, Shay B Cohen, and Mirella Lapata. 2018. Ranking sentences for extractive summarization with reinforcement learning. In NAACL-HLT, pages 1747–1759.
- Nguyen et al. (2017) Gia-Hung Nguyen, Laure Soulier, Lynda Tamine, and Nathalie Bricon-Souf. 2017. Dsrim: A deep neural information retrieval model enhanced by a knowledge resource driven representation of documents. In Proceedings of the ACM SIGIR International Conference on Theory of Information Retrieval, pages 19–26.
- Nogueira and Cho (2017) Rodrigo Nogueira and Kyunghyun Cho. 2017. Task-oriented query reformulation with reinforcement learning. In Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, pages 574–583.
- Noh and Kavuluru (2018) Jiho Noh and Ramakanth Kavuluru. 2018. Document retrieval for biomedical question answering with neural sentence matching. In 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA), pages 194–201. IEEE.
- Paulus et al. (2018) Romain Paulus, Caiming Xiong, and Richard Socher. 2018. A deep reinforced model for abstractive summarization. International Conference on Learning Representations.
- Prasad and Kan (2019) Animesh Prasad and Min-Yen Kan. 2019. Glocal: Incorporating global information in local convolution for keyphrase extraction. In NAACL-HLT, pages 1837–1846.
- Rios and Kavuluru (2019) Anthony Rios and Ramakanth Kavuluru. 2019. Neural transfer learning for assigning diagnosis codes to emrs. Artificial Intelligence in Medicine, 96:116–122.
- Rios et al. (2018) Anthony Rios, Ramakanth Kavuluru, and Zhiyong Lu. 2018. Generalizing biomedical relation classification with neural adversarial domain adaptation. Bioinformatics, 34(17):2973–2981.
- Roberts et al. (2020) Kirk Roberts, Dina Demner-Fushman, Ellen M. Voorhees, William R. Hersh, Steven Bedrick, Alexander J. Lazar, Shubham Pant, and Funda Meric-Bernstam. 2020. Overview of the TREC 2019 precision medicine track.
- Roberts et al. (2016) Kirk Roberts, Matthew Simpson, Dina Demner-Fushman, Ellen Voorhees, and William Hersh. 2016. State-of-the-art in biomedical literature retrieval for clinical cases: a survey of the trec 2014 cds track. Information Retrieval Journal, 19(1):113–148.
- Rush et al. (2015) Alexander M Rush, Sumit Chopra, and Jason Weston. 2015. A neural attention model for abstractive sentence summarization. In EMNLP, pages 379–389.
- Schuster and Nakajima (2012) Mike Schuster and Kaisuke Nakajima. 2012. Japanese and korean voice search. In 2012 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pages 5149–5152. IEEE.
- See et al. (2017) Abigail See, Peter J Liu, and Christopher D Manning. 2017. Get to the point: Summarization with pointer-generator networks. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics, pages 1073–1083.
- Sun et al. (2019) Zhiqing Sun, Jian Tang, Pan Du, Zhi-Hong Deng, and Jian-Yun Nie. 2019. Divgraphpointer: A graph pointer network for extracting diverse keyphrases. In Proceedings of the 42nd International ACM SIGIR Conference on Research and Development in Information Retrieval, pages 755–764.
- Vaswani et al. (2017) Ashish Vaswani, Noam Shazeer, Niki Parmar, Jakob Uszkoreit, Llion Jones, Aidan N Gomez, Łukasz Kaiser, and Illia Polosukhin. 2017. Attention is all you need. In Advances in neural information processing systems, pages 5998–6008.
- Wagner et al. (2020) Alex H Wagner, Brian Walsh, Georgia Mayfield, David Tamborero, Dmitriy Sonkin, Kilannin Krysiak, Jordi Deu-Pons, Ryan P Duren, Jianjiong Gao, Julie McMurry, et al. 2020. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nature genetics, 52(4):448–457.
- Wolf et al. (2019) Thomas Wolf, Lysandre Debut, Victor Sanh, Julien Chaumond, Clement Delangue, Anthony Moi, Pierric Cistac, Tim Rault, Rémi Louf, Morgan Funtowicz, et al. 2019. Transformers: State-of-the-art natural language processing. arXiv preprint arXiv:1910.03771.
- Wu et al. (2016) Yonghui Wu, Mike Schuster, Zhifeng Chen, Quoc V Le, Mohammad Norouzi, Wolfgang Macherey, Maxim Krikun, Yuan Cao, Qin Gao, Klaus Macherey, et al. 2016. Google’s neural machine translation system: Bridging the gap between human and machine translation. arXiv preprint arXiv:1609.08144.
Appendix A Attention Heatmaps by Facet Signals
Figure 3 depicts words highlighted by EXT. Evidently, we see terms related to the regulations of gene expressions, proteins, or disease names featuring more prominently. Figure 4 shows how ABS reads the source document differently depending on which facet signal it starts with, in the process of query generation; compared to [unused0] (disease facet), the attention heat map by [unused1] (genetic facet) focuses more on the words related to gene regulations.


