Is it Time to Replace CNNs with Transformers for Medical Images?
Abstract
Convolutional Neural Networks (CNNs) have reigned for a decade as the de facto approach to automated medical image diagnosis. Recently, vision transformers (ViTs) have appeared as a competitive alternative to CNNs, yielding similar levels of performance while possessing several interesting properties that could prove beneficial for medical imaging tasks. In this work, we explore whether it is time to move to transformer-based models or if we should keep working with CNNs – can we trivially switch to transformers? If so, what are the advantages and drawbacks of switching to ViTs for medical image diagnosis? We consider these questions in a series of experiments on three mainstream medical image datasets. Our findings show that, while CNNs perform better when trained from scratch, off-the-shelf vision transformers using default hyperparameters are on par with CNNs when pretrained on ImageNet, and outperform their CNN counterparts when pretrained using self-supervision.
1 Introduction
††Originally published at the ICCV 2021 Workshop on Computer Vision for Automated Medical Diagnosis (CVAMD).
Vision transformers have gained increased popularity for image recognition tasks recently, signalling a transition from convolution-based feature extractors (CNNs) to attention-based models (ViTs). Following the success of Dosovitskiy et al. [11], numerous approaches for adapting transformers to vision tasks have been suggested [17]. In the natural image domain, transformers have been shown to outperform CNNs on standard vision tasks such as ImageNet classification, [11] as well as in object detection [3] and semantic segmentation [21]. The attention mechanism central to transformers offers several key advantages over convolutions: (1) it captures long-range relationships, (2) it has the capacity for adaptive modeling via dynamically computed self-attention weights that capture relationships between tokens, (3) it provides a type of built-in saliency which gives insight as to what the model focused on [4].
Yet, evidence suggests that vision transformers require very large datasets to outperform CNNs – in [11], the benefits of ViT only became evident when Google’s private 300 million image dataset, JFT-300M, was used for pretraining. Their reliance on data of this scale is a barrier to the widespread application of transformers. This problem is particularly acute in the medical imaging domain, where datasets are smaller and are often accompanied by less reliable labels.
CNNs, like ViTs, suffer worse performance when data is scarce. The standard solution is to employ transfer learning: typically, a model is pretrained on a larger dataset such as ImageNet [10] and then fine-tuned for specific tasks using smaller, specialized, datasets. CNNs pre-trained on ImageNet typically outperform those trained from scratch in the medical domain, both in terms of final performance and reduced training time [20].
Self-supervision is a learning approach to deal with unlabeled data that has recently gained much attention. It has been shown that self-supervised pretraining of CNNs in the target domain before fine-tuning can increase performance [1]. Initialization from ImageNet helps self-supervised CNNs converge faster, and usually with better predictive performance [1].
These techniques to deal with the lack of data in the medical image domain have proven effective for CNNs, but it remains unclear whether vision transformers benefit similarly. Some studies suggest that pre-training CNNs for medical image analysis using ImageNet does not rely on feature reuse –following conventional wisdom– but, rather due to better initialization and weight scaling [20]. This calls into question whether transformers benefit from these techniques. If they do, there is little to prevent ViTs from becoming the dominant architecture for medical images.
In this work, we explore whether ViTs can easily replace CNNs for medical imaging tasks, and if there is an advantage of doing so. We consider the use-case of a typical practitioner, equipped with a limited computational budget and access to conventional medical datasets, with an eye towards “plug-and-play” solutions. To this end, we conduct experiments on three mainstream publicly available datasets. Through these experiments we show that:
-
•
ViTs pretrained on ImageNet perform comparably to CNNs when data is limited.
-
•
Transfer learning favours ViTs when applying standard training protocols and settings.
-
•
ViTs outperform their CNN counterparts when self-supervised pre-training is followed by supervised fine-tuning.
These findings suggest that medical image analysis can seamlessly transition from CNNs to ViTs, while at the same time gaining improved explainability properties. To promote transparency and reproducibility, we share our open-source code, available at https://github.com/ChrisMats/medical_transformers.
2 Related Work
The use of vision transformers in the natural imaging domain has exploded recently, with applications ranging from classification [11], to object detection [3] and segmentation [21]. In medical imaging, however, the use of ViTs has been limited – primarily focused on focused on segmentation [6, 19]. Only a handful of studies have tackled other tasks such as 3-D image registration [5] and detection [12]. Notably, none of these works consider pure, off-the-shelf, vision transformers – all propose custom architectures combining transformer/attention modules with convolutional feature extractors.
Although it is well-known that CNNs usually benefit from transfer learning to medical imaging domains, the source of these benefits is disputed. The conventional wisdom that feature re-use contributes to better performance was questioned by Raghu et al. [20], who rather attribute the improved performance to good initialization and weight statistics. Regardless of the reason, the question of whether ViTs benefit from transfer learning to medical domains is yet to be explored.
Recent advances in self-supervised learning have dramatically improved performance of label-free learning. State-of-the-art methods such as DINO [4] and BYOL [13] have reached performance on par with supervised learning on ImageNet and other standard benchmarks. While these top-performing methods have not yet been proven for medical imaging, Azizi et al. [1] adopted SimCLR [7], an earlier self-supervised contrastive learning method, to pretrain CNNs. This yielded state-of-the-art results for predictions on chest X-rays and skin lesions. However, it has yet to be shown how self-supervised learning combined with ViTs performs in medical imaging, and whether this combination outperforms its CNN counterparts.
| Initialization | Model | APTOS2019, | ISIC2019, Recall | DDSM, ROC-AUC |
|---|---|---|---|---|
| Random | ResNet50 | 0.849 0.022 | 0.662 0.018 | 0.917 0.005 |
| DeiT-S | 0.687 0.017 | 0.579 0.028 | 0.908 0.015 | |
| ImageNet (supervised) | ResNet50 | 0.893 0.004 | 0.810 0.008 | 0.953 0.008 |
| DeiT-S | 0.896 0.005 | 0.844 0.021 | 0.947 0.011 | |
| ImageNet (supervised) + Self-supervised with DINO [4] | ResNet50 | 0.894 0.008 | 0.833 0.007 | 0.955 0.002 |
| DeiT-S | 0.896 0.010 | 0.853 0.009 | 0.956 0.002 |
3 Methods
The main question we investigate is whether ViTs can be used as a plug-and-play alternative to CNNs for medical diagnostic tasks. To that end, we conducted a series of experiments to compare vanilla ViTs and CNNs under similar conditions, keeping hyperparameter tuning to a minimum. To ensure a fair and interpretable comparison, we selected ResNet50 [15] as the representative CNN model, and DeiT-S with tokens [23] as the ViT. These models were chosen because they are comparable in the number of parameters, memory requirements, and compute.
As mentioned above, CNNs rely on initialization strategies to improve performance when data is less abundant, as is the case for medical images. The standard approach is to use transfer learning – initialize the model with weights pretrained on ImageNet and fine-tune on the target domain. More recently, self-supervised pretraining has become a popular way to initialize neural networks [13, 4, 1].
Accordingly, we consider three initialization strategies: (1) randomly initialized weights, (2) transfer learning using supervised ImageNet pretrained weights, (3) self-supervised pretraining on the target dataset, after initialization as in (2). We apply these strategies on three standard medical imaging datasets chosen to cover a diverse set of target domains:
-
•
APTOS 2019 – In this dataset, the task is classification of diabetic retinopathy images into 5 categories of disease severity [16]. APTOS 2019 contains 3,662 high-resolution retinal images.
- •
-
•
CBIS-DDSM – This dataset contains 10,239 mammography images and the task is to detect the presence of masses in the mammograms.
Datasets were divided into train/test/validation splits (80/10/10), with the exception of APTOS, which was divided 70/15/15 due to its small size. All supervised training uses an Adam optimizer [18] with a base learning rate of with a warm-up period of 1,000 iterations. When the validation metrics saturate, the learning rate is dropped by a factor of 10 until it reaches its final value of . All images are resized to and standard augmentations were applied111Training augmentations include: normalization; color jitter including brightness, contrast, saturation, hue; horizontal flip; vertical flip; and random resized crops. We repeat each experiment five times, and select the checkpoint with highest validation score of each run. We use the above settings unless otherwise specified.
4 Experiments
Are randomly initialized transformers useful?
We begin by comparing DeiT-S against ResNet50 with randomly initialized weights (Kaiming initialization [14]). For these experiments, the base learning rate was set to 0.0003 following a grid search. The results in Table 1 indicate that in this setting, CNNs outperform ViTs by a large margin across the board. These results are in line with previous observations in the natural image domain, where ViTs trained on limited data are outperformed by similarly-sized CNNs, a trend that was attributed to ViT’s lack of inductive bias [11]. Since most medical imaging datasets are of modest size, the usefulness of randomly initialized ViTs appears to be limited.
Does pretraining transformers on ImageNet work in the medical image domain?
On medical image datasets, random initialization is rarely used in practice. The standard procedure is to train CNNs by initializing the network with ImageNet pretrained weights, followed by fine-tuning on data from the target domain. Here, we investigate if this approach can be effectively applied to vanilla ViTs. To test this, we initialize all models with weights that have been pre-trained on ImageNet in a fully-supervised fashion. We then fine-tune using the procedure described above. The results in Table 1 show that both CNNs and ViTs benefit significantly from ImageNet initialization. In fact, ViTs appear to benefit more, as they perform on par with their CNN counterparts. This indicates that, when initialized with ImageNet, CNNs can be replaced with vanilla ViTs without compromising performance on medical imaging tasks with modest-sized training data.
Do transformers benefit from self-supervision in the medical image domain?
Recent self-supervised learning schemes such as DINO [4] and BYOL [13] approach supervised learning performance. Moreover, if they are used for pretraining in combination with supervised fine-tuning, they can achieve a new state-of-the-art [4, 13]. While this phenomenon has been demonstrated for CNNs and ViTs in larger data regimes, it is not clear whether self-supervised pretraining of ViTs helps for medical imaging tasks, especially on modest- and low-sized data. To test this, we adopt the self-supervised learning scheme of DINO [4], which can be readily applied to both CNNs and ViTs. DINO uses self-distillation to encourage a student and teacher network to produce similar representations given differently-augmented inputs. Our self-supervised pretraining starts with ImageNet initialization, then applies self-supervised learning on the target medical domain data following the default settings suggested by the authors of the original paper [4] – except for three small changes: (1) the base learning rate was set to 0.0001, (2) the initial weight decay is set at and increased to using a cosine schedule, and (3) we used an EMA of 0.99. The same settings were used for both CNNs and ViTs; both were pre-trained for 300 epochs using a batch size of 256, followed by fine-tuning as described in Section 3.
5 Discussion
Our investigation compares the performance of vanilla CNNs and ViTs on medical image tasks under three different initialization strategies. The results of the experiments corroborate previous findings and provide new insights.
In medical images, as was previously reported in the natural image domain, we found that CNNs outperform ViTs when trained from scratch in a low data regime [11]. This trend appears consistently across all the datasets and fits well with the “transformers lack inductive bias” argument.
Surprisingly, when initialized with supervised ImageNet pretrained weights, the gap between CNN and ViT performance disappears on medical tasks. The benefits of supervised ImageNet pretraining on CNNs is well-known, but it was unexpected that ViTs would benefit so strongly. To the best of our knowledge, we are the first to confirm that supervised ImageNet pretraining is as effective for ViTs as it is for CNNs on medical imaging tasks. This suggests that further improvements could be gained via transfer learning from other domains more closely related to the task, as is the case for CNNs [2].
We investigated the effect of self-supervised pre-training on the medical image domain. Our results indicate small but consistent improvements for ViTs and CNNs. While the best overall performance is obtained using self-supervised ViTs, interestingly in this low-data regime we do not yet see the strong advantage for self-supervision favoring ViTs that was previously reported in the natural image domain with more data, e.g. in [4]
Large labeled medical image datasets are rare due to the cost of expert annotation, but it may be possible to gather large amounts of unlabeled images. This suggests a tantalizing opportunity to apply self-supervision on large medical image datasets, where only a small fraction are labeled.
To summarize our findings, for the medical image domain:
-
•
As expected, ViTs are worse than CNNs in the low data regime if one simply trains from scratch.
-
•
Transfer learning bridges the performance gap between CNNs and ViTs; performance is similar.
-
•
The best performance is obtained with self-supervised pre-training + fine-tuning, where ViTs enjoy a small advantage over comparable CNNs.
Interpretability.
It appears that ViTs can replace CNNs for medical image tasks – are there any other reasons for choosing ViTs over CNNs? One should consider the added benefits of visualizing transformer attention maps. Built-in to the self-attention mechanism of transformers is an attention map that provides, for free, new insight into how the model makes decisions. CNNs do not naturally lend themselves well to visualizing saliency. Popular CNN explainability methods such as class activation maps (CAM) [25] and Grad-CAM [22] provide coarse visualizations because of pooled layers. Transformer tokens give a finer picture of attention, and the self-attention maps explicitly model interactions between every region in the image, in contrast to the limited receptive field of the CNN. While the difference in quality of explainability has yet to be quantified, many have noted the qualitative improvements in interpretability afforded by transformer attention [4].
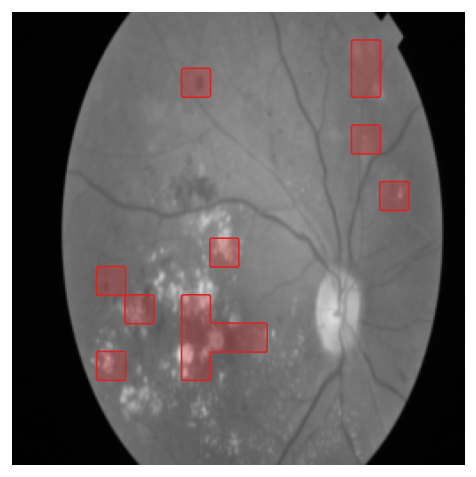
In Figure 1 we show examples from each of the datasets, along with Grad-CAM visualizations of the ResNet-50 and the top-50% self-attention of DeiT-S CLS token heads. Notice how the self-attention of ViTs provide a clear, localized picture of the attention, e.g. attention at the boundary of the skin lesion in ISIC, on hemorrhages and exudates in APTOS, and at the dense region of the breast in CBIS-DDSM. This granularity of attention is difficult to achieve with CNNs.
 |
 |
 |
 |
 |
 |
 |
 |
 |
| ISIC 2019 | APTOS 2019 | CBIS-DDSM |
6 Conclusion
Finally, to answer the question posed in the title: vanilla transformers can reliably replace CNNs on medical image tasks with little effort. More precisely, ViTs can reach the same level of performance as CNNs in small medical datasets, but require transfer learning in order to do so. However, using ImageNet pretrained weights is the standard approach for CNNs as well, so the switch to ViTs is trivial. Furthermore, ViTs can outperform CNNs using SSL pre-training when working with limited number of samples, but only marginally. As the number of samples grows, the margin between ViT and CNN is expected to grow as well. This equal or better performance comes with the additional benefit of built in high-resolution saliency maps that can be used to better understand the model’s decisions.
Acknowledgements.
This work was supported by the Wallenberg Autonomous Systems Program (WASP), and the Swedish Research Council (VR) 2017-04609. We thank Moein Sorkhei and Emir Konuk for the thoughtful discussions.
References
- [1] Shekoofeh Azizi, Basil Mustafa, Fiona Ryan, Zachary Beaver, Jan Freyberg, Jonathan Deaton, Aaron Loh, Alan Karthikesalingam, Simon Kornblith, Ting Chen, et al. Big self-supervised models advance medical image classification. arXiv preprint arXiv:2101.05224, 2021.
- [2] Hossein Azizpour, Ali Sharif Razavian, Josephine Sullivan, Atsuto Maki, and Stefan Carlsson. Factors of transferability for a generic convnet representation. IEEE transactions on pattern analysis and machine intelligence, 38(9):1790–1802, 2016.
- [3] Nicolas Carion, Francisco Massa, Gabriel Synnaeve, Nicolas Usunier, Alexander Kirillov, and Sergey Zagoruyko. End-to-end object detection with transformers. In European Conference on Computer Vision, pages 213–229. Springer, 2020.
- [4] Mathilde Caron, Hugo Touvron, Ishan Misra, Hervé Jégou, Julien Mairal, Piotr Bojanowski, and Armand Joulin. Emerging properties in self-supervised vision transformers. arXiv preprint arXiv:2104.14294, 2021.
- [5] Junyu Chen, Yufan He, Eric C Frey, Ye Li, and Yong Du. Vit-v-net: Vision transformer for unsupervised volumetric medical image registration. arXiv preprint arXiv:2104.06468, 2021.
- [6] Jieneng Chen, Yongyi Lu, Qihang Yu, Xiangde Luo, Ehsan Adeli, Yan Wang, Le Lu, Alan L Yuille, and Yuyin Zhou. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306, 2021.
- [7] Ting Chen, Simon Kornblith, Mohammad Norouzi, and Geoffrey Hinton. A simple framework for contrastive learning of visual representations. In International conference on machine learning, pages 1597–1607. PMLR, 2020.
- [8] Noel CF Codella, David Gutman, M Emre Celebi, Brian Helba, Michael A Marchetti, Stephen W Dusza, Aadi Kalloo, Konstantinos Liopyris, Nabin Mishra, Harald Kittler, et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018), pages 168–172. IEEE, 2018.
- [9] Marc Combalia, Noel CF Codella, Veronica Rotemberg, Brian Helba, Veronica Vilaplana, Ofer Reiter, Cristina Carrera, Alicia Barreiro, Allan C Halpern, Susana Puig, et al. Bcn20000: Dermoscopic lesions in the wild. arXiv preprint arXiv:1908.02288, 2019.
- [10] J. Deng, W. Dong, R. Socher, L.-J. Li, K. Li, and L. Fei-Fei. ImageNet: A Large-Scale Hierarchical Image Database. In CVPR09, 2009.
- [11] Alexey Dosovitskiy, Lucas Beyer, Alexander Kolesnikov, Dirk Weissenborn, Xiaohua Zhai, Thomas Unterthiner, Mostafa Dehghani, Matthias Minderer, Georg Heigold, Sylvain Gelly, et al. An image is worth 16x16 words: Transformers for image recognition at scale. In International Conference on Learning Representations, 2020.
- [12] Linh T Duong, Nhi H Le, Toan B Tran, Vuong M Ngo, and Phuong T Nguyen. Detection of tuberculosis from chest x-ray images: Boosting the performance with vision transformer and transfer learning. Expert Systems with Applications, page 115519, 2021.
- [13] Jean-Bastien Grill, Florian Strub, Florent Altché, Corentin Tallec, Pierre H Richemond, Elena Buchatskaya, Carl Doersch, Bernardo Avila Pires, Zhaohan Daniel Guo, Mohammad Gheshlaghi Azar, et al. Bootstrap your own latent: A new approach to self-supervised learning. arXiv preprint arXiv:2006.07733, 2020.
- [14] Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE international conference on computer vision, pages 1026–1034, 2015.
- [15] Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. Deep residual learning for image recognition. In The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 2016.
- [16] Kaggle. Aptos 2019 blindness detection, 2019.
- [17] Salman Khan, Muzammal Naseer, Munawar Hayat, Syed Waqas Zamir, Fahad Shahbaz Khan, and Mubarak Shah. Transformers in vision: A survey. arXiv preprint arXiv:2101.01169, 2021.
- [18] Diederik P Kingma and Jimmy Ba. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
- [19] Daniel H Pak, Andrés Caballero, Wei Sun, and James S Duncan. Efficient aortic valve multilabel segmentation using a spatial transformer network. In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), pages 1738–1742. IEEE, 2020.
- [20] Maithra Raghu, Chiyuan Zhang, Jon Kleinberg, and Samy Bengio. Transfusion: Understanding transfer learning for medical imaging. In Advances in Neural Information Processing Systems, pages 3342–3352, 2019.
- [21] René Ranftl, Alexey Bochkovskiy, and Vladlen Koltun. Vision transformers for dense prediction. arXiv preprint arXiv:2103.13413, 2021.
- [22] Ramprasaath R Selvaraju, Michael Cogswell, Abhishek Das, Ramakrishna Vedantam, Devi Parikh, and Dhruv Batra. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE international conference on computer vision, pages 618–626, 2017.
- [23] Hugo Touvron, Matthieu Cord, Matthijs Douze, Francisco Massa, Alexandre Sablayrolles, and Hervé Jégou. Training data-efficient image transformers & distillation through attention. In International Conference on Machine Learning, pages 10347–10357. PMLR, 2021.
- [24] Philipp Tschandl, Cliff Rosendahl, and Harald Kittler. The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data, 5(1):1–9, 2018.
- [25] Bolei Zhou, Aditya Khosla, Agata Lapedriza, Aude Oliva, and Antonio Torralba. Learning deep features for discriminative localization. In Proceedings of the IEEE conference on computer vision and pattern recognition, pages 2921–2929, 2016.