Ghent University Global Campus, Republic of Korea 22institutetext: Department of Applied Mathematics, Computer Science and Statistics,
Ghent University, Belgium 33institutetext: Department of Electronics and Information Systems,
Ghent University, Belgium
33email: [email protected]
Identifying Critical Tokens for
Accurate Predictions in
Transformer-based Medical Imaging Models
Abstract
With the advancements in self-supervised learning (SSL), transformer-based computer vision models have recently demonstrated superior results compared to convolutional neural networks (CNNs) and are poised to dominate the field of artificial intelligence (AI)-based medical imaging in the upcoming years. Nevertheless, similar to CNNs, unveiling the decision-making process of transformer-based models remains a challenge. In this work, we take a step towards demystifying the decision-making process of transformer-based medical imaging models and propose \sayToken Insight, a novel method that identifies the critical tokens that contribute to the prediction made by the model. Our method relies on the principled approach of token discarding native to transformer-based models, requires no additional module, and can be applied to any transformer model. Using the proposed approach, we quantify the importance of each token based on its contribution to the prediction and enable a more nuanced understanding of the model’s decisions. Our experimental results which are showcased on the problem of colonic polyp identification using both supervised and self-supervised pretrained vision transformers indicate that Token Insight contributes to a more transparent and interpretable transformer-based medical imaging model, fostering trust and facilitating broader adoption in clinical settings.
1 Introduction
While pretrained CNNs [12, 24] have been the go-to models for solving complex medical imaging problems in the past decade, the emergence of transformer-based architectures such as the vision transformer (ViT) [8] and the self-attention mechanism [27] originating from natural language processing (NLP) has significantly influenced the computer vision community, marking a shift towards replacing CNNs as the preferred models. The emergence of foundational models as well as other advances in SSL frameworks such as DINO [4], MAE [11], MoCo [7], and many more [17] has further sped up the adoption rate of transformer-based models. Nowadays, such models are employed in medical imaging to solve a variety of medical-imaging problems, with recent studies suggesting that ViTs generally outperform CNNs [16, 23, 25].
Unfortunately, similar to other types of neural networks, transformer-based networks also struggle in in reasoning capabilities [1, 22]. That is, it is not straightforward to decipher the reason for predictions that have been made by transformer networks. To address the issue of lack of transparency for transformers, a number of novel interpretability methods for ViTs have been proposed, with some of these efforts making use of techniques borrowed from research on CNNs [5], while others focus on new areas native to transformers, such as the usage of attention maps and token-based methods [20, 2, 30]. These interpretability methods often produce what are called interpretability maps, with the goal of highlighting the salient regions of the image. In the context of medical imaging, these regions often correspond to areas where the disease occurs (for example, highlighting a tumor). As a result, the performance of such methods is often evaluated based on their ability to highlight medically-relevant parts that are annotated by experts. However, recent research on spurious correlations suggests that the salient parts of the medical images identified by models might not be the same as the ones identified by experts [26]. Indeed, certain undesired signals may lead to what is called shortcut learning and result in models with undesired behaviors that perform the task at hand via signals that are not intended [9].

To address these challenges with predictive reasoning, how can we identify the parts of medical images that lead to the predictions the model has made, regardless of undesired model behaviors such as shortcut learning? This work is centered around answering this question. Our method, called \sayToken Insight, identifies the critical tokens that contribute to the predictions made by a ViT. This method relies on the principled approach of token discarding in ViTs, which has garnered significant interest for several reasons in recent years, for example to speed up training [3, 14], to create robust models [19], as well as to provide interpretability [10]. In contrast to most interpretability methods that use a variety of metrics as a proxy for predictive reasoning (such as the overlap with salient regions), our method directly uses the reduction in prediction confidence as a means to identify critical tokens. As a result, Token Insight provides a more transparent and intuitive understanding of the model’s decision-making process, leading to the identification of undesired model behaviors such as shortcut learning, offering a more reliable means to pinpoint the specific aspects of tokens influencing the model’s predictions.
2 Methodology
In this section, we provide a description of the dataset used, outline the models utilized, and finally describe the proposed method in detail.
| Training | Testing | Device | |||
|---|---|---|---|---|---|
| Dataset | Non-polyp | Polyp | Non-polyp | Polyp | |
| CP-CHILD-A | 6,200 | 800 | 800 | 200 | Olympus PCF-H290DI |
| CP-CHILD-B | 800 | 300 | 300 | 100 | FUJIFILM EC-530wm |
| Combined | 7,000 | 1,100 | 1,100 | 300 | Both |
2.1 Dataset
For straightforward and easily understandable experiments, we utilize the combination of recently proposed CP-CHILD-A and CP-CHILD-B datasets, which involves the detection of colonic polyps [28]. Both datasets combined comprises of 9,500 colonoscopy RGB images obtained from 1,600 patients using the Olympus PCF-H290DI and FUJIFILM EC-530wm. The task involves identifying colonic polyps, which is challenging since it is not trivial to distinguish polyps from other problematic colonic tissues such as lesions, ulcerative colitis, and inflammatory bowel disease. Furthermore, many of the polyps presented in the dataset do not appear fully in the picture and are only visible in the corners, thus increasing the challenge. Further details regarding data splits of these datasets are provided in Table 1.
2.2 Models
To showcase the capabilities of the proposed method, we employ the most commonly used transformer-based computer vision architecture, Vision Transformer-Base/16 (ViT-B/16) [8]. As suggested by [8], we use this model for images of size with image tokens of size , resulting in a total of tokens. We modify the final linear layer of the model to accommodate the two-class classification problem tackled in this work.
We use two ViT-B/16 models where the first one is randomly initialized and trained from scratch while the second one is pretrained in a supervised fashion. Apart from those two models, in order to capture various properties of self-supervised learning methods that have seen increased use in medical imaging problems in recent years, we employ two additional models pretrained using (1) DINO [4], a discriminative SSL framework, and (2) MAE [11], which relies on a generative approach. The pretraining for the aforementioned models is performed on the ImageNet dataset [21], a large-scale dataset containing natural images.
2.3 Token Insight
Given an image and its categorical association , sampled from a dataset , where and for all , let represent a vision transformer with parameters that maps an image to a set of prediction likelihoods, denoted as , where . If , then the classification is considered correct.
When utilized as an input for a vision transformer, the image X undergoes a process of patchification (also called tokenization), transforming it into a set of tokens as shown in Figure 1. In this study, we adopt ViT-B/16 which employs patches. Given that the input images have a resolution of , the input X is patchified into tokens denoted as . As shown in Figure 1, in the setup of ViT, a special token called [cls] token is prepended on , thus increasing the number of tokens by one. This token is then used for the purpose of making classification using a linear layer after the final transformer encoder layer.
Token Insight is designed to progressively identify the most critical token contributing to the model’s prediction. To achieve this, at each step, it removes one token at a time, measures the change in prediction, and identifies the token causing the largest drop in prediction confidence for the correct class. Subsequently, after examining each token, the one resulting in the highest confidence change when removed is permanently discarded. This process is then repeated to discover remaining critical tokens until the prediction ultimately changes. In the case of polyp detection problem, Token Insight finds the tokens that lead to the prediction of the \sayPolyp class and the token discarding operation is carried out repeatedly until the prediction changes from \sayPolyp-positive to \sayPolyp-negative for the images considered. Token Insight finds its origins in the works of [15, 18]. However, unlike those approaches which stop after a single iteration, we continue to search for critical tokens greedily and discard them until the prediction is eventually changed, thus identifying all of the critical tokens that contribute to the prediction.
Formally, we can represent Token Insight as follows: given an input image represented as , denote by the image where the tokens have been removed. At the next iteration, , the most critical token is the token that gives rise to the largest drop in prediction confidence:
The iterative search described above stops when . Note that Token Insight does not alter or remove the [cls] token during the process described above and, in order to generate the prediction confidences [cls] token is used according to the description of [8] throughout the process.
An illustration of the token discarding operation of Token Insight is provided in Figure 2 where the example image is initially predicted with confidence as having a polyp. Using Token Insight, critical tokens are identified and discarded iteratively until the prediction eventually drops down to for the polyp-positive class, thus identifying tokens that contribute to the prediction made by the model.
Relation to occlusion-based methods – Note that, on the surface, Token Insight appears to be similar to the occlusion-based methods which mask certain regions of the input to measure the change in the prediction [29]. The primary difference between Token Insight and these methods is that during the token discarding process the model is not affected by the potential spurious signals such as missingness bias introduced by the masking operation (i.e., the bias that is introduced by the mask) [13]. Since the tokens are completely removed, the model prediction is not influenced by the removed tokens. Unfortunately, since the token removal operation is only available for transformer-based models, Token Insight is not usable for CNNs and other non-transformer DNN architectures.
3 Experimental Results
3.1 Training
As described in Section 2.2 and Section 2.1, we employ four ViT-B/16 models, three of which are pretrained on the ImageNet dataset, and train them on the combined CP-Child dataset. To discover the most appropriate model with the highest performance, we perform extensive training efforts and in what follows, we detail the training routine that leads to the identification of the best model.
For pretrained models (supervised, DINO, MAE), training is performed for epochs using Stochastic Gradient Descent (SGD) with an initial learning rate of and momentum of . The model trained from scratch undergoes a longer training period ( epochs) to compensate for the lack of pretraining and uses an increased learning rate of , weight decay of , and momentum of . All models employ a batch size of and use cosine annealing, which reduces the learning rate after each epoch using the method described in [6].
In the work of [28], pretrained ResNets are reported to attain an accuracy of approximately on the CP-Child dataset. With pretrained ViTs, we replicate these results, achieving comparable performance. The model trained from scratch exhibits slightly reduced performance at due to the absence of pretraining. Nonetheless, this model still delivers commendable results on this dataset, making these models suitable for a study on predictive reasoning.





3.2 Token Insight
In order to investigate various properties of the proposed approach as well as models pretrained in different ways, we apply Token Insight to the images containing polyps in the CP-Child-B dataset.
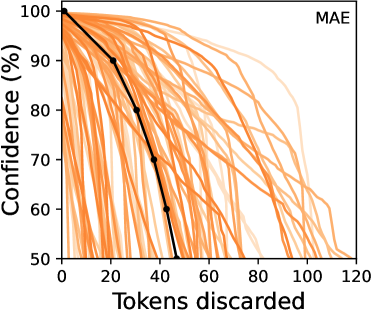
Number of tokens discarded – For all four models, we investigate the number of tokens discarded via Token Insight to examine the model’s reliance on tokens for making a polyp-positive prediction. Doing so, in Figure 3(b), we present the number of tokens discarded to change a polyp-positive prediction into a polyp-negative one. As it can be seen, both MAE and the model trained from scratch discard more tokens before the prediction changes, while models pretrained with supervised learning and DINO discard fewer, and hence rely on more tokens to make a prediction. This implies that treating all transformer-based medical AI models the same would be a mistake since the number of tokens used for confirming a prediction may vary significantly depending on how the model is trained.
The most impactful token – Expanding our investigation, in Figure 3(b), we illustrate the influence of the most impactful tokens across the dataset, as measured by their impact on the prediction confidence when removed. Complementing our previous insights, we observe that individual tokens in the models pretrained with supervised learning, as well as DINO, have more influence on the prediction compared to MAE and the model trained from scratch. Once again, this observation reveals that it is possible to have two models sharing the same architecture, but where one heavily relies on fewer tokens, whereas the other focuses on a broader area. Consequently, depending on the medical imaging task at hand, practitioners may prefer one type of model over another.
Combining the two experiments discussed above, we present Figure 3(c) which shows confidence drops per removed token over the number of tokens discarded for each model. Graphs presented in Figure 3(c) can be seen as a summarizing views of Figure 3(b) and Figure 3(b), where the goal is to reveal the different behaviors of various models. As can be seen, all models exhibit an initial stage in which confidence drops only moderately as the initial few tokens are removed, followed by a steep drop in confidence. MAE and scratch see a relative robustness in the decrease of prediction confidence as a function of the number of tokens removed, compared to DINO and supervised pretrained models.
Identifying shortcut learning – In Figure 4, we present several examples showcasing the application of Token Insight on images containing polyps. Note that, while Token Insight accurately identifies regions containing polyps, thereby indicating that the model learns the correct signals, there are instances (particularly for MAE and Scratch) where certain tokens are identified as important but are not medically relevant to the prediction. This suggests that the model under consideration is indeed making predictions based on signals that are not intended in the dataset, also identified by [26]. As demonstrated, the primary use-case of the proposed approach is to uncover such cases and to discover predictions based on erroneous signals, thereby enhancing the trustworthiness of AI-based medical imaging.
4 Conclusion
In this work, we introduced Token Insight, a novel computational method designed to identify critical tokens that contribute to the prediction for transformer-based medical imaging models, thus enabling its usage as a method for identifying spurious correlations as well as erroneous signals that lead to shortcut learning. Unlike many of its predecessors, this method does not necessitate any changes in the model or require any additional extra modules. Furthermore, the results obtained by the proposed approach rely entirely on the change in prediction confidence of the model, making it a reliable indicator for identifying the undesired cases mentioned above.
We foresee two possible directions for future work. The current method identifies a set of tokens that influence the prediction by greedily removing the highest impact token at every iteration, and we have offered computational evidence that the set thus obtained forms a good approximation to the smallest set of highly influential tokens. For the future, we plan to investigate theoretical bounds for how well our algorithm manages to capture the smallest set of tokens that influence the prediction. A second line of work concerns the computational cost of our method, which scales as in the number of tokens. Given the considerable computational cost involved in a forward pass of a transformer-based model, it would be of interest to re-use previously acquired information about the impact of a token to avoid making a forward pass for each candidate token.
5 Acknowledgements
This work was supported by a grant from the Special Research Fund (BOF) of Ghent University (BOF/STA/202109/039).
References
- [1] Bai, B., Liang, J., Zhang, G., Li, H., Bai, K., Wang, F.: Why attentions may not be interpretable? In: Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining. pp. 25–34 (2021)
- [2] Bastings, J., Filippova, K.: The elephant in the interpretability room: Why use attention as explanation when we have saliency methods? arXiv preprint arXiv:2010.05607 (2020)
- [3] Bolya, D., Fu, C.Y., Dai, X., Zhang, P., Feichtenhofer, C., Hoffman, J.: Token merging: Your vit but faster. arXiv preprint arXiv:2210.09461 (2022)
- [4] Caron, M., Touvron, H., Misra, I., Jégou, H., Mairal, J., Bojanowski, P., Joulin, A.: Emerging properties in self-supervised vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 9650–9660 (2021)
- [5] Chefer, H., Gur, S., Wolf, L.: Transformer interpretability beyond attention visualization. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 782–791 (2021)
- [6] Chen, X., He, K.: Exploring simple siamese representation learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 15750–15758 (2021)
- [7] Chen, X., Xie, S., He, K.: An empirical study of training self-supervised vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 9640–9649 (2021)
- [8] Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., et al.: An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
- [9] Geirhos, R., Jacobsen, J.H., Michaelis, C., Zemel, R., Brendel, W., Bethge, M., Wichmann, F.A.: Shortcut learning in deep neural networks. Nature Machine Intelligence 2(11), 665–673 (2020)
- [10] Haurum, J.B., Escalera, S., Taylor, G.W., Moeslund, T.B.: Which tokens to use? investigating token reduction in vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 773–783 (2023)
- [11] He, K., Chen, X., Xie, S., Li, Y., Dollár, P., Girshick, R.: Masked autoencoders are scalable vision learners. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 16000–16009 (2022)
- [12] He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (2016)
- [13] Jain, S., Salman, H., Wong, E., Zhang, P., Vineet, V., Vemprala, S., Madry, A.: Missingness bias in model debugging. arXiv preprint arXiv:2204.08945 (2022)
- [14] Long, S., Zhao, Z., Pi, J., Wang, S., Wang, J.: Beyond attentive tokens: Incorporating token importance and diversity for efficient vision transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 10334–10343 (2023)
- [15] Madsen, A., Reddy, S., Chandar, S.: Faithfulness measurable masked language models. arXiv preprint arXiv:2310.07819 (2023)
- [16] Matsoukas, C., Haslum, J.F., Söderberg, M., Smith, K.: Is it time to replace cnns with transformers for medical images? arXiv preprint arXiv:2108.09038 (2021)
- [17] Ozbulak, U., Lee, H.J., Boga, B., Anzaku, E.T., Park, H., Van Messem, A., De Neve, W., Vankerschaver, J.: Know your self-supervised learning: A survey on image-based generative and discriminative training. arXiv preprint arXiv:2305.13689 (2023)
- [18] Pan, B., Panda, R., Jiang, Y., Wang, Z., Feris, R., Oliva, A.: : Interpretability-aware redundancy reduction for vision transformers. Advances in Neural Information Processing Systems 34, 24898–24911 (2021)
- [19] Renggli, C., Pinto, A.S., Houlsby, N., Mustafa, B., Puigcerver, J., Riquelme, C.: Learning to merge tokens in vision transformers. arXiv preprint arXiv:2202.12015 (2022)
- [20] Rigotti, M., Miksovic, C., Giurgiu, I., Gschwind, T., Scotton, P.: Attention-based interpretability with concept transformers. In: International Conference on Learning Representations (2021)
- [21] Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., Huang, Z., Karpathy, A., Khosla, A., Bernstein, M., Berg, A.C., Fei-Fei, L.: ImageNet large scale visual recognition challenge. International Journal of Computer Vision 115(3), 211–252 (2015)
- [22] Serrano, S., Smith, N.A.: Is attention interpretable? arXiv preprint arXiv:1906.03731 (2019)
- [23] Shamshad, F., Khan, S., Zamir, S.W., Khan, M.H., Hayat, M., Khan, F.S., Fu, H.: Transformers in medical imaging: A survey. Medical Image Analysis p. 102802 (2023)
- [24] Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. International Conference on Learning Representations (2015)
- [25] Singhal, K., Tu, T., Gottweis, J., Sayres, R., Wulczyn, E., Hou, L., Clark, K., Pfohl, S., Cole-Lewis, H., Neal, D., et al.: Towards expert-level medical question answering with large language models. arXiv preprint arXiv:2305.09617 (2023)
- [26] Sun, S., Koch, L.M., Baumgartner, C.F.: Right for the wrong reason: Can interpretable ml techniques detect spurious correlations? In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 425–434. Springer (2023)
- [27] Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., Polosukhin, I.: Attention is all you need. Advances in neural information processing systems 30 (2017)
- [28] Wang, W., Tian, J., Zhang, C., Luo, Y., Wang, X., Li, J.: An improved deep learning approach and its applications on colonic polyp images detection. BMC Medical Imaging 20, 1–14 (2020)
- [29] Zeiler, M.D., Fergus, R.: Visualizing and understanding convolutional networks. In: Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6-12, 2014, Proceedings, Part I 13. pp. 818–833. Springer (2014)
- [30] Zeng, W., Jin, S., Liu, W., Qian, C., Luo, P., Ouyang, W., Wang, X.: Not all tokens are equal: Human-centric visual analysis via token clustering transformer. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 11101–11111 (2022)