Gaussian RBF Centered Kernel Alignment (CKA) in the Large Bandwidth Limit
Abstract
We prove that Centered Kernel Alignment (CKA) based on a Gaussian RBF kernel converges to linear CKA in the large-bandwidth limit. We show that convergence onset is sensitive to the geometry of the feature representations, and that representation eccentricity bounds the range of bandwidths for which Gaussian CKA behaves nonlinearly.
Index Terms:
Nonlinear kernels, neural networks, representations, similarity.1 Introduction
Centered Kernel Alignment (CKA) was first proposed as a measure of similarity between kernels in the context of kernel learning [1, 2], building on prior work on (non-centered) kernel-target alignment [3]. Versions for functional data of CKA and the associated Hilbert-Schmidt Independence Criterion (HSIC) were proposed later [4]. CKA has been used for multiple kernel learning (e.g., [5], [6]). CKA also enables measuring similarity between two feature representations of a set of data examples (e.g., two sets of neural network activation vectors), by comparing kernel-based similarity matrices (Gram matrices) of these representations [7]. Used in the latter manner, CKA and alternatives such as canonical correlation analysis (CCA) and orthogonal Procrustes distance [8] can provide insight into the relationship between architectural features of a network such as width and depth, on one hand, and the network’s learned representations [7, 9], on the other. CKA has been used to assess representation similarity in cross-lingual language models [10] and recurrent neural network dynamics [11], to identify potential drug side-effects [12], to differentiate among brain activity patterns [13], and to gauge the representational effects of watermarking [14], among others.
Any positive-definite symmetric kernel can be used as the base kernel for CKA, including linear, polynomial, and Gaussian radial basis function (RBF) kernels. Gaussian RBF kernels are of special interest due to their universality properties [15], which can lead to superior modeling. Understanding the behavior of CKA with Gaussian RBF kernels is, therefore, relevant to its use as a representational similarity metric. Some prior work [7] reports finding little difference empirically between CKA similarity values based on Gaussian RBF and linear kernels. In contrast, hyperparameter tuning experiments for [16] suggest that a noticeable difference can occur between CKA values for Gaussian RBF and linear kernels, but also that the difference becomes negligible as Gaussian bandwidth grows. We establish the latter phenomenon theoretically in the present paper, proving that Gaussian CKA converges to linear CKA as bandwidth approaches infinity, for all representations. We describe the convergence rate, as well.
We are not aware of previously published results on the large-bandwidth asymptotics of Gaussian CKA. A limit result for Gaussian kernel SVM classification appeared in [17], but relies on an analysis of the dual formulation of the maximum-margin optimization problem that does not translate directly to kernel CKA.
While we find that approximation of Gaussian RBF CKA by linear CKA for large bandwidths becomes apparent by elementary means, our proof shows that the result relies on centering of the feature maps. Indeed, we show that a similar result does not hold for the analogous non-centered kernel-target alignment measure of [3]. We also show that a geometric measure of representation eccentricity serves to bound the range of bandwidths for which Gaussian and linear CKA differ for a given representation. This can be helpful in data-adaptive Gaussian bandwidth selection.
2 Background and notation
Our perspective is that of measuring similarity between two feature representations of the same set of data examples. We briefly review the basic ingredients and notation related to kernel similarity and CKA below (see [7], [2]).
2.1 Feature representations
and will denote matrices of -dimensional (resp., -dimensional) feature vectors for the same set of data examples. Each row of or consists of the feature vector for one of these examples. We use subindices to indicate the rows of and (e.g., , ).
2.2 Kernel similarity
Linear similarity between feature encodings , can be expressed as similarity of their “similarity structures” (within-encoding dot product matrices) , [7]:
| (1) |
where is the Frobenius norm and is the trace function. The left-hand side reflects similarity of the feature representations; the right-hand side reflects similarity of their respective self-similarity matrices.
CKA (defined in section 2.3) corresponds to a normalized version of Eq. 1, extended to kernel similarity by replacing and by Gram matrices and for positive-definite symmetric kernel functions, and . The bars indicate that columns have been mean-centered [18], [2]; see section 2.5.
Gram matrix entries can be viewed as inner products of the embedded images of the data examples in a high-dimensional reproducing kernel Hilbert space (RKHS) [19]. Thus, kernels allow modeling aspects of representations not easily accessible to the linear version.
2.3 HSIC and CKA
The kernel similarity perspective applied to Eq. 1 yields the Hilbert-Schmidt Independence Criterion (HSIC) [18] shown in Eq. 2, where denotes the number of data examples, and the dependence on and has been hidden for economy of notation. Mean-centering of the Gram matrices is assumed to have been carried out as described in section 2.5, below.
| (2) |
CKA (Eq. 3) is a normalized version of HSIC; it takes values in the interval , as observed in [2].
| (3) |
2.4 Kernels and Gram matrices
We consider Gram matrices and , where and denote positive-definite (p.d.) symmetric kernel functions. We focus on linear and Gaussian RBF kernels; the Euclidean (polynomial, degree ) kernel is also used. See Eq. LABEL:eq:kernelOptions, where is dot product and is Euclidean norm. Gram matrices have size , regardless of the dimensionality of the feature representation.
| Linear | |||||
| Euclidean (quadratic) |
Note on scaling of distances in Gaussian Gram matrices
Following the heuristic of [7], we use instead of in Eq. LABEL:eq:kernelOptions when computing the Gram matrix , where is the median distance between examples (rows) of (e.g., describes a bandwidth equal to twice the median distance between examples). Equivalently, we divide all inter-example distances by . This ensures invariance of Gaussian HSIC under isotropic scaling [7].
2.5 Mean-centering
Mean-centering the Gram matrices in Eq. 2 and Eq. 3 is crucial both for kernel learning [2] and for the results in this paper. Mean-centering of the Gram matrices corresponds to centering the embedded features in the RKHS. Centering in the embedding space appeared in early work on kernel PCA [20].
We default to column mean-centering, that is, we ensure that each column has mean zero. Thus, we multiply by the centering matrix, , on the left as in Eq. 5a (where is the identity matrix and is an matrix of ones), with individual entries as in Eq. 5b. For row mean-centering, would multiply from the right and the summation would range over columns instead of rows.
| (5a) | |||
| (5b) | |||
Our results hold for mean-centering of either rows or columns, and for simultaneous centering of both. The latter case [2] involves multiplication by on both sides in Eq. 5a; two terms are added to the right in Eq. 5b, corresponding to subtracting times the column-sum of the current Eq. 5b.
3 Convergence proof and derivation of a bandwidth-selection heuristic
We prove convergence of Gaussian CKA to linear CKA for large bandwidths in section 3.1, and provide a data-dependent criterion that bounds the range of Gaussian bandwidths for which nonlinear behavior occurs, in Section 3.2.
3.1 Linear CKA approximation of Gaussian RBF CKA
Our main result is Theorem 1. We focus on the case in which only one of the two Gram matrix parameters uses a Gaussian RBF kernel with bandwidth , as doing so leads to cleaner proofs; the other, , is assumed to be any fixed, positive-definite symmetric kernel. The result holds equally if both Gram matrices are Gaussian with bandwidths approaching infinity (see Appendix in Supplemental Material).
Theorem 1.
as . In particular, converges to as . The result also holds if both kernels are Gaussian RBF kernels; in that case, the limit as both bandwidths approach infinity is (i.e., it is ).
We prove Theorem 1 by showing first, in Lemma 1, that Gaussian CKA converges to Euclidean CKA, and then, as a Corollary to Lemma 2, that Euclidean CKA and linear CKA are identical.
Note
Our results rely crucially on mean-centering of features in Eqs. 2, 3. Indeed, the analog of Theorem 1 does not hold if, as in [3], features are not centered. This is easy to see by a direct calculation for the example in which feature matrix is the identity and feature matrix is a of ones, except for a single off-diagonal . Linear CKA equals in that case, while Gaussian CKA is for all , as and have identical distance matrices after scaling by the median distances , as described in the note in section 2.4. Details are provided in the Appendix.
Lemma 1.
as , for any positive-definite kernel, .
Proof of Lemma 1: Let . Also, let , where is the median pairwise distance between feature vectors in . Then, by Eq. LABEL:eq:kernelOptions and Eq. 5b, the centered Gram matrix has the entries shown in Eq. 6. Mean-centering is an indispensable ingredient, as we will see.
| (6) |
The argument uses the first-order series expansion of the exponential function at the origin (Eq. 7).
| (7) |
Applying Eq. 7 to the exponential terms of Eq. 6 as , the importance of mean-centering becomes apparent. Due to subtraction of the sum for the column mean in Eq. 6, the constant terms in Eq. 7 cancel, leaving only the centered Euclidean Gram matrix entries and higher-order terms:
The form of the residual term persists in the HSIC expression of Eq. 2:
Multiplying by , we obtain the connection in Eq. 8 between and :
| (8) | ||||
An identity analogous to Eq. 8 relates to its Euclidean version. In the latter case, the higher-order error terms in the HSIC expression are before multiplying by , because they result from a product of a error term from one of the two Gaussian Gram matrices with the leading term in the other factor; multiplication by is required to obtain a residual, which will become a factor after taking the square root in the CKA denominator (below).
Lemma 1 follows by dividing the HSIC components described above to form CKA as in Eq. 3:
| (9) | ||||
The case of two Gaussian kernels with bandwidths approaching (whether equal to one another or not) follows by considering one kernel argument at a time, freezing the bandwidth of the other, and using the scalar triangle inequality. Details appear in the Appendix.
Lemma 2.
, for any p.d. .
Proof: First, we show that the column-centered Gram matrix entries can be written as , where the term depends only on the row, , not the column, :
The quantity in parentheses is , and the remaining term depends only on , as stated. We can now relate the HSIC expressions:
Since the columns of have mean zero, the sum on the right is zero. If row-centering is used instead of column-centering, the conclusion follows by first expressing the entries of the Euclidean Gram matrix as times the linear Gram matrix entries plus a term that depends only on the column.
Corollary 1.
for any kernel .
3.2 Bounding the nonlinear Gaussian bandwidth range
A heuristic bound on the term in Theorem 1 follows along the same lines as the proof of Lemma 1, by examining how the residual grows as the CKA terms are assembled, and how its magnitude is constrained by geometric characteristics of the feature representations. We consider this issue in the case in which a Gaussian kernel is used for both feature maps, as this potentially maximizes the residual due to contributions from both Gram matrix factors in Lemma 1. We show, below, how convergence onset reflects the representation-specific range of variation of pairwise distances between data examples in feature space.
The power series for in Eq. 7 is alternating if , with decreasing terms after the second if ; the error term in Eq. 7 is then no larger than . Relative error with respect to the linear term is . Multiplying or dividing two terms roughly doubles relative error, since and ; likewise, since , taking square roots halves relative error.
Let ; then each of the exponential arguments in the Gram matrices in the proof of Lemma 1 is bounded by . It follows by the error considerations in the preceding paragraph that HSIC relative error in Eq. 8 is approximately bounded by , and CKA relative error in Eq. 9 is bounded by . Notice that , where the “representation eccentricity”, , is defined as in Eq. 10 (so depends only on pairwise distances, and , with equality only if all pairwise distances are equal).
| (10) |
Thus, CKA relative error is approximately bounded by , so that Gaussian RBF CKA approximately equals Euclidean CKA for bandwidths . For lower-dimensional representations , , the representation eccentricity, , provides a data-sensitive approximate threshold between nonlinear and linear regimes for Gaussian CKA. If feature dimensionality is high, concentration of Euclidean distance [21] will make ; hence, for high-dimensional representations, one expects that Gaussian CKA will behave linearly if .
4 Experimental illustration
In this section we describe the results of a limited number of experiments that compare CKA similarity of neural feature representations based on Gaussian kernels of different bandwidths, with linear CKA similarity. For simplicity, we restrict attention to the case in which both of the kernels in Eq. 3 are of the same type, either Gaussian RBF kernels of equal bandwidth, or standard linear kernels. Software for these experiments is available from the author upon request.
Notation
In this section, we write as shorthand for . Likewise, we abbreviate (i.e., ) as .
4.1 Experimental setup
Data sets
We used sample OpenML data sets [22] (CC BY 4.0 license, https://creativecommons.org/licenses/by/4.0/) for classification (splice, tic-tac-toe, wdbc, optdigits, wine, dna) and regression (cpu, boston, Diabetes(scikit-learn), stock, balloon, cloud); for data sets with multiple versions, we used version 1. These data sets were selected based on two considerations only: smaller size, in order to eliminate the need for GPU acceleration and reduce environmental impact; and an absence of missing values, to simplify the development of internal feature representations for the CKA computations. Data set sizes are given in Table I.
| Classification | Regression | |||||
|---|---|---|---|---|---|---|
| data set | examples | attributes | data set | examples | attributes | |
| splice | 3190 | 60 | cpu | 209 | 7 | |
| tic-tac-toe | 958 | 9 | boston | 506 | 13 | |
| wdbc | 569 | 30 | Diabetes(scikit-learn) | 442 | 10 | |
| optdigits | 5620 | 64 | stock | 950 | 9 | |
| wine | 178 | 13 | balloon | 2001 | 1 | |
| dna | 3186 | 180 | cloud | 108 | 5 | |
Neural feature representations
Fully-connected neural networks (NN) with two hidden layers were used. Feature encodings , were the sets of activation vectors of the first and second hidden layers, respectively. Alternative NN widths, were tested, with and hidden nodes in the first and second hidden layers. A configuration with three hidden layers, of sizes , was also tested, for which , were the activation vectors from layers and .
Implementation
NN were trained using the MLPClassifier and MLPRegressor classes in scikit-learn [23], with cross-entropy or quadratic loss for classification and regression, respectively, ReLU activation functions, Glorot-He pseudorandom initialization [24], [25], Adam optimizer [26], learning rate of , and a maximum of training iterations. CKA was implemented in Python (https://docs.python.org/3/license.html), using NumPy [27] (https://numpy.org/doc/stable/license.html) and Matplotlib [28] (PSF license, https://docs.python.org/3/license.html). Experiments were performed on a workstation with an Intel i9-7920X (12 core) processor and 128GB RAM, under Ubuntu 18.04.5 LTS (GNU public license).
Experiments
runs were performed for each pair of a data set and NN width . In each run, a new NN model was trained on the full data set, starting from fresh pseudorandom initial parameter values. A training run was repeated if the resulting in-sample accuracy was below (classification) or if in-sample coefficient of determination was below (regression), but not if the optimization had not converged within the allowed number of iterations. After the network had been trained in a given run, was computed once, and was computed for each bandwidth , for a total of and evaluations per pair.
Compute time
Compute time for the results presented in the paper was approximately hours, much of it on the CKA evaluations needed across the network widths (including the 128-32-8 three-hidden-layer configuration) and the data sets. Additional runs for validation required another hours. Several shorter preliminary runs were carried out for debugging and initial selection of the NN hyperparameters; configurations and threshold values were selected empirically in order to ensure a successful end to training after no more than a handful of runs in nearly all cases. Total compute time across all runs is estimated to have been hours.
Evaluation metrics
CKA means and standard errors (SE = standard deviation divided by ), and medians and standard error equivalent (SE = inter-quartile range divided by ) of the ratio of Eq. 10 were computed across runs. We measured the magnitude of the discrepancy between and by the base- logarithm of the relative difference between the two measures as in Eq. 11.
| (11) |
Theorem 1 implies that, for large , the logarithmic relative difference of Eq. 11 should decrease along a straight line of slope as a function of , reflecting a dependence. Accordingly, for each data set, we determined a asymptote by extrapolating backward from the largest tested bandwidth value, . If , the predicted relative difference along the asymptote is as in Eq. 12, where is the observed relative difference at .
| (12) |
We determined a convergence onset bandwidth, , as the minimum bandwidth above which the observed relative difference between Gaussian and linear CKA differs by less than from the predicted value along that data set’s asymptote (Eq. 13).
| (13) |
Threshold values other than in Eq. 13 (), corresponding to different tolerances for the log relative difference, yielded similar results in most cases in terms of the resulting relative ranks of the different data sets.
4.2 Discussion of experimental results
of maximum to median distance between features. is NN width. Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| 16 | 10.63 0.53 | 5.08 0.54 | 4.87 0.01 | 3.09 0.28 | 14.62 0.88 | 6.59 0.45 |
| 32 | 9.60 0.58 | 3.87 0.23 | 4.86 0.01 | 2.68 0.14 | 15.64 0.83 | 5.76 0.38 |
| 64 | 9.23 0.29 | 3.57 0.29 | 4.86 0.01 | 2.62 0.12 | 16.25 0.90 | 5.99 0.26 |
| 128 | 9.15 0.20 | 3.61 0.11 | 4.85 0.01 | 2.50 0.05 | 18.49 0.86 | 5.84 0.19 |
| 256 | 9.19 0.16 | 3.40 0.07 | 4.84 0.01 | 2.48 0.05 | 20.70 0.88 | 6.05 0.10 |
| 512 | 8.91 0.12 | 3.38 0.04 | 4.84 0.01 | 2.44 0.04 | 22.65 0.72 | 6.20 0.12 |
| 1024 | 8.82 0.11 | 3.42 0.03 | 4.82 0.04 | 2.43 0.02 | 25.18 1.14 | 6.16 0.19 |
| 128-32-8 | 9.96 0.54 | 3.91 0.24 | 4.83 0.01 | 3.11 0.25 | 22.71 2.97 | 7.30 0.23 |
The results show a range of geometric characteristics of learned feature representations across data sets, seen as differences in the representation eccentricity, , defined in Eq. 10. Geometry is stable for a given data set, as indicated by a small standard deviation for , with some dependence on network size. See Table II for the case of regression; the Appendix includes the information for classification.
We observe generally lower values of (Eq. 10) for wider networks in Table II. This is consistent with expectations, as concentration of Euclidean distance in high dimensions [21] will bring the ratio of maximum to median distance between feature vectors closer to as network width grows. The balloon data set is the only one for which increases with network width. That data set contains a small group of examples with lower values of the two attributes than the rest (indices 332, 706, 969, 1025, 1041, 1399, 1453, 1510). Those examples are increasingly distant from the majority (in units of median distance) in the deeper (second or third) hidden layer representations as width increases, driving the increase in ; the ratio of maximum to median distance in the first hidden layer representation does not exhibit similar growth.
Dimensionality of the raw data sets (Table I) appears to filter into the learned neural representations, as well. For a fixed network width, the correlation between raw data dimensionality and is unambiguously negative, between and for classification, and between and for regression, even though representation dimensionality is fixed by network width.
Noticeable differences between Gaussian and linear CKA occur for small bandwidths
Mean relative difference values between and greater than ( values greater than ), are observed for several data sets when (when ) in the case of classification; in fact, relative CKA difference values greater than (, values in the Appendix) are observed for the dna data set when network width is or greater. This shows that noticeably nonlinear behavior of Gaussian CKA is quite possible for small bandwidth values. For some classification data sets, however (wdbc, wine), and most regression data sets, the relative CKA difference remains comparatively small for all bandwidths.
Gaussian CKA converges to linear CKA like
Fig. 1 shows confidence intervals for the means, of radius two standard errors, of the observed relative difference between and as a function of Gaussian bandwidth, (Eq. 11). Convergence at the rate for is observed (log-log slope of in Fig. 1), as described in Theorem 1. Results are stable across the range of neural network configurations tested. Standard error of the relative CKA difference is observed to decrease as network width increases (e.g., Fig. 2), suggesting that wider networks are less sensitive to variations in initial parameter values.

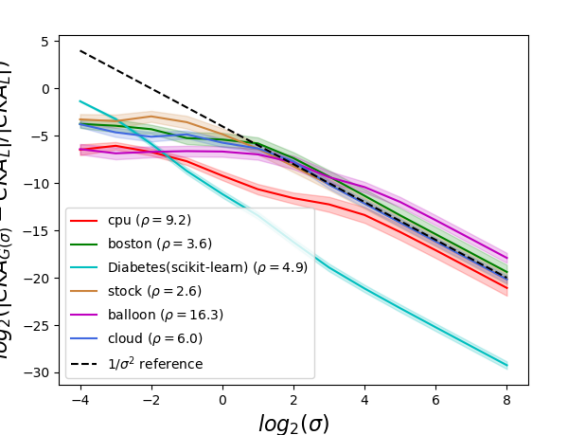


The Diabetes(scikit-learn) data set stands out for having, by far, the smallest mean relative CKA difference among data sets tested, across much of the range. The neural feature maps and for that data set have nearly proportional linear Gram matrices and , hence the linear CKA value is very close to . Because the matrix of squared inter-example distances depends linearly on the linear Gram matrix, the Gaussian CKA value is also very close to .
The representation eccentricity (Eq. 10) is reflected in convergence onset
Our experimental results suggest that noticeably nonlinear behavior of Gaussian CKA occurs almost exclusively for bandwidths . Indeed, for all data sets tested, the observed mean relative difference between linear and Gaussian CKA is less than when , where is the ratio of maximum to median distance between feature vectors (Eq. 10). This confirms our finding in section 3.2 that differs little from if . Furthermore, our results suggest that the threshold between nonlinear and linear regimes of Gaussian CKA is not substantially less than : while mean relative CKA difference for peaked under for two-hidden-layer neural representations, across all data sets tested, relative difference values less than occur for the tic-tac-toe data set in the three-layer 128-32-8 representation only when ; the lower end of this bandwidth range is quite close to the median value of for that representation (which differs from the width- two-layer representation in Figs. 1, 2).
We find additional support for the view that approximates the boundary between nonlinear and linear regimes of Gaussian CKA in the fact that the convergence onset bandwidth, (Eq. 13), is largest for the data sets of largest : median , , for balloon (), wdbc (), cpu (), respectively ( and values differ slightly among network widths and depths, but data sets of largest are the same). See Table III. Diabetes(scikit-learn) ties for third with ; median for all other data sets. Large differences in between data sets generally translate directly to differences between their values. This is consistent with the analysis in section 3.2, as also controls terms of orders higher than in the exponential power series.
Our experimental results as a whole confirm convergence of to as (Theorem 1), and support the use of the representation eccentricity, (Eq. 10), as a useful heuristic upper bound on the range of bandwidths for which Gaussian CKA can display nonlinear behavior.
| Classification | Regression | ||||||||||||
| splice | t-t-t | wdbc | optd | wine | dna | cpu | bost | diab | stock | balln | cloud | ||
| 16 | 2 | 2 | 5 | 2 | -1 | 2 | 4 | 3 | 4 | 1 | 5 | 4 | |
| 32 | 2 | 2 | 4 | 1 | 3 | 2 | 4 | 3 | 4 | 1 | 5 | 4 | |
| 64 | 2 | 2 | 4 | 1 | 3 | 3 | 5 | 2 | 4 | 2 | 5 | 3 | |
| 128 | 3 | 1 | 4 | 1 | 2 | 2 | 4 | 2 | 4 | 1 | 5 | 2 | |
| 256 | 3 | 1 | 4 | 1 | 3 | 2 | 5 | 2 | 4 | 1 | 5 | 3 | |
| 512 | 3 | 1 | 4 | 1 | 3 | 2 | 4 | 2 | 4 | 1 | 5 | 2 | |
| 1024 | 2 | 1 | 4 | 1 | 3 | 2 | 5 | 2 | 5 | 1 | 5 | 2 | |
| 128-32-8 | 3 | 2 | 4 | 1 | 2 | 2 | 4 | 2 | 3 | 0 | 6 | 3 | |
5 Conclusions
This paper considered the large-bandwidth asymptotics of CKA using Gaussian RBF kernels. We proved rigorously that mean-centering of the feature vectors ensures that Gaussian RBF CKA converges to linear CKA in the large bandwidth limit, with an asymptotic relative difference between them. We showed that a similar result does not hold for the non-centered kernel alignment measure of [3].
We also showed that the geometry of the feature representations impacts the range of bandwidths for which Gaussian CKA can behave nonlinearly, focusing on the representation eccentricity ratio, , of maximum to median distance between feature vectors as a representation-sensitive guide. Our results suggest that bandwidth values less than can lead to noticeably nonlinear behavior of Gaussian CKA, whereas bandwidths larger than will yield essentially linear behavior. In order to enable nonlinear modeling, the bandwidth should, therefore, be selected in the interval .
6 Future Work
Representation eccentricity, , correlates well with the bandwidth at which Gaussian CKA transitions between nonlinear and linear regimes, and our theoretical result establishes convergence of Gaussian to linear CKA for large bandwidths. In applications, however, the order of magnitude of the difference between Gaussian and linear CKA can be of greatest interest in regard to kernel selection. One direction for future work would be to seek additional representation characteristics that, in conjunction with eccentricity, can better gauge the order of magnitude of the Gaussian-linear CKA difference for a given representation. Such work could provide further guidance in selecting between Gaussian and linear CKA kernels for particular applications. Robust versions of the representation eccentricity can also be explored, in which maximum and median distance are replaced by other pairs of quantiles of the distance distribution.
Declarations
The work for this paper did not involve human subjects. Data sets used do not include any personally identifiable information or offensive content. The author has no conflicts of interest to report.
Acknowledgments
The author thanks Sarun Paisarnsrisomsuk, whose experiments for [16] motivated this paper, and Carolina Ruiz, for helpful comments on an earlier version of the manuscript.
References
- [1] C. Cortes, M. Mohri, and A. Rostamizadeh, “Two-stage learning kernel algorithms,” in Proc. 27th Intl. Conf. on Machine Learning, ser. ICML’10. Madison, WI, USA: Omnipress, 2010, p. 239–246.
- [2] ——, “Algorithms for learning kernels based on centered alignment,” J. Mach. Learn. Res., vol. 13, no. 28, pp. 795–828, 2012. [Online]. Available: http://jmlr.org/papers/v13/cortes12a.html
- [3] N. Cristianini, J. Shawe-Taylor, A. Elisseeff, and J. Kandola, “On kernel-target alignment,” in Adv. in Neur. Inf. Proc. Systems, T. Dietterich, S. Becker, and Z. Ghahramani, Eds., vol. 14. MIT Press, 2002. [Online]. Available: https://proceedings.neurips.cc/paper/2001/file/1f71e393b3809197ed66df836fe833e5-Paper.pdf
- [4] T. Górecki, M. Krzyśko, and W. Wołyński, “Independence test and canonical correlation analysis based on the alignment between kernel matrices for multivariate functional data,” Artif Intell Rev, vol. 53, p. 475–499, 2020. [Online]. Available: https://doi.org/10.1007/s10462-018-9666-7
- [5] Y. Lu, L. Wang, J. Lu, J. Yang, and C. Shen, “Multiple kernel clustering based on centered kernel alignment,” Pattern Recognition, vol. 47, no. 11, pp. 3656–3664, 2014. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S0031320314001903
- [6] S. Niazmardi, A. Safari, and S. Homayouni, “A novel multiple kernel learning framework for multiple feature classification,” IEEE J. Sel. Topics Appl. Earth Obs. and Remote Sensing, vol. 10, no. 8, pp. 3734–3743, 2017.
- [7] S. Kornblith, M. Norouzi, H. Lee, and G. E. Hinton, “Similarity of neural network representations revisited,” in Proc. 36th Intl. Conf. Mach. Learn., ICML 2019, 9-15 June 2019, Long Beach, California, USA, ser. Proc. Mach. Learn. Res., K. Chaudhuri and R. Salakhutdinov, Eds., vol. 97. PMLR, 2019, pp. 3519–3529. [Online]. Available: http://proceedings.mlr.press/v97/kornblith19a.html
- [8] F. Ding, J.-S. Denain, and J. Steinhardt, “Grounding representation similarity through statistical testing,” in Thirty-Fifth Conference on Neural Information Processing Systems (NeurIPS), 2021. [Online]. Available: https://openreview.net/forum?id=_kwj6V53ZqB
- [9] T. Nguyen, M. Raghu, and S. Kornblith, “Do Wide and Deep Networks Learn the Same Things? Uncovering How Neural Network Representations Vary with Width and Depth,” CoRR, vol. abs/2010.15327, 2020 (ICLR 2021). [Online]. Available: https://arxiv.org/abs/2010.15327
- [10] A. Conneau, S. Wu, H. Li, L. Zettlemoyer, and V. Stoyanov, “Emerging cross-lingual structure in pretrained language models,” in Proc. 58th Ann. Meeting Assoc. Comp. Ling. Online: Assoc. Comp. Ling., Jul. 2020, pp. 6022–6034. [Online]. Available: https://www.aclweb.org/anthology/2020.acl-main.536
- [11] N. Maheswaranathan, A. Williams, M. Golub, S. Ganguli, and D. Sussillo, “Universality and individuality in neural dynamics across large populations of recurrent networks,” in Adv. Neur. Inf. Proc. Syst., H. Wallach, H. Larochelle, A. Beygelzimer, F. d'Alché-Buc, E. Fox, and R. Garnett, Eds., vol. 32. Curran Associates, Inc., 2019. [Online]. Available: https://proceedings.neurips.cc/paper/2019/file/5f5d472067f77b5c88f69f1bcfda1e08-Paper.pdf
- [12] Y. Ding, J. Tang, and F. Guo, “Identification of drug-side effect association via multiple information integration with centered kernel alignment,” Neurocomputing, vol. 325, pp. 211–224, 2019. [Online]. Available: https://www.sciencedirect.com/science/article/pii/S0925231218312165
- [13] A. M. Alvarez-Meza, A. Orozco-Gutierrez, and G. Castellanos-Dominguez, “Kernel-based relevance analysis with enhanced interpretability for detection of brain activity patterns,” Frontiers in Neuroscience, vol. 11, p. 550, 2017. [Online]. Available: https://www.frontiersin.org/article/10.3389/fnins.2017.00550
- [14] H. Jia, C. A. Choquette-Choo, V. Chandrasekaran, and N. Papernot, “Entangled watermarks as a defense against model extraction,” in 30th USENIX Security Symposium (USENIX Security 21). USENIX Association, Aug. 2021. [Online]. Available: https://www.usenix.org/conference/usenixsecurity21/presentation/jia
- [15] B. K. Sriperumbudur, K. Fukumizu, and G. R. Lanckriet, “Universality, characteristic kernels and RKHS embedding of measures,” J. Mach. Learn. Res., vol. 12, no. 70, pp. 2389–2410, 2011. [Online]. Available: http://jmlr.org/papers/v12/sriperumbudur11a.html
- [16] S. Paisarnsrisomsuk, “Understanding internal feature development in deep convolutional neural networks for time series,” Ph.D. dissertation, Worcester Polytechnic Institute, Aug. 2021.
- [17] S. S. Keerthi and C.-J. Lin, “Asymptotic behaviors of support vector machines with Gaussian kernel,” Neur. Comp., vol. 15, no. 7, pp. 1667–1689, 2003.
- [18] A. Gretton, O. Bousquet, A. Smola, and B. Schölkopf, “Measuring statistical dependence with Hilbert-Schmidt norms,” in Proceedings of the 16th International Conference on Algorithmic Learning Theory, ser. ALT’05. Berlin, Heidelberg: Springer-Verlag, 2005, p. 63–77. [Online]. Available: https://doi.org/10.1007/11564089_7
- [19] T. Hofmann, B. Schölkopf, and A. J. Smola, “Kernel methods in machine learning,” Ann. Stat., vol. 36, no. 3, pp. 1171–1220, 2008.
- [20] B. Schölkopf, A. Smola, and K.-R. Müller, “Nonlinear Component Analysis as a Kernel Eigenvalue Problem,” Neural Computation, vol. 10, no. 5, pp. 1299–1319, 07 1998. [Online]. Available: https://doi.org/10.1162/089976698300017467
- [21] C. C. Aggarwal, A. Hinneburg, and D. A. Keim, “On the surprising behavior of distance metrics in high dimensional space,” in Database Theory — ICDT 2001, J. Van den Bussche and V. Vianu, Eds. Berlin, Heidelberg: Springer Berlin Heidelberg, 2001, pp. 420–434.
- [22] J. Vanschoren, J. N. van Rijn, B. Bischl, and L. Torgo, “OpenML: Networked science in machine learning,” SIGKDD Explorations, vol. 15, no. 2, pp. 49–60, 2013. [Online]. Available: http://doi.acm.org/10.1145/2641190.2641198
- [23] F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss, V. Dubourg, J. Vanderplas, A. Passos, D. Cournapeau, M. Brucher, M. Perrot, and E. Duchesnay, “Scikit-learn: Machine learning in Python,” J. Machine Learning Research, vol. 12, pp. 2825–2830, 2011.
- [24] X. Glorot and Y. Bengio, “Understanding the difficulty of training deep feedforward neural networks,” in Proc. 13th Intl. Conf. AI and Stat. (AISTATS 2010), Chia Laguna Resort, Sardinia, Italy, May 13-15, 2010, ser. JMLR Proceedings, Y. W. Teh and D. M. Titterington, Eds., vol. 9. JMLR.org, 2010, pp. 249–256. [Online]. Available: http://proceedings.mlr.press/v9/glorot10a.html
- [25] K. He, X. Zhang, S. Ren, and J. Sun, “Delving deep into rectifiers: Surpassing human-level performance on imagenet classification,” in Proc. 2015 IEEE Intl. Conf. Comp. Vis. (ICCV), ser. ICCV ’15. USA: IEEE Computer Society, 2015, p. 1026–1034. [Online]. Available: https://doi.org/10.1109/ICCV.2015.123
- [26] D. P. Kingma and J. Ba, “Adam: A method for stochastic optimization,” in 3rd Intl Conf. Learn. Repr. (ICLR 2015), San Diego, CA, USA, May 7-9, 2015, Conf. Track Proc., Y. Bengio and Y. LeCun, Eds., 2015. [Online]. Available: http://arxiv.org/abs/1412.6980
- [27] C. R. Harris, K. J. Millman, S. J. van der Walt, R. Gommers, P. Virtanen, D. Cournapeau, E. Wieser, J. Taylor, S. Berg, N. J. Smith, R. Kern, M. Picus, S. Hoyer, M. H. van Kerkwijk, M. Brett, A. Haldane, J. F. del Río, M. Wiebe, P. Peterson, P. Gérard-Marchant, K. Sheppard, T. Reddy, W. Weckesser, H. Abbasi, C. Gohlke, and T. E. Oliphant, “Array programming with NumPy,” Nature, vol. 585, no. 7825, pp. 357–362, Sep. 2020. [Online]. Available: https://doi.org/10.1038/s41586-020-2649-2
- [28] J. D. Hunter, “Matplotlib: A 2D graphics environment,” Computing in Science & Engineering, vol. 9, no. 3, pp. 90–95, 2007.
Appendix A Supplementary information for the paper
Gaussian RBF CKA
in the large bandwidth limit
A.1 Details of the counterexample for non-centered kernel alignment in section 3.1
As we state in section 3.1, Theorem 1 hinges on the assumption that the Gram matrices are mean-centered. We justify that statement here. We show that an analogous convergence result does not hold for the non-centered kernel alignment of [3], by considering the feature matrices below.
In order to compute , we first compute, for each of the feature matrices and , the corresponding matrix of distances between pairs of feature vectors (rows):
and are scalings of one another. Given a bandwidth, , we interpret as expressed in units of median distance, as in the note in section 2.4. If diagonal zeros are included in the median computation, then in the non-centered Gaussian entries of , and in the non-centered Gaussian entries of . Median scaling makes the non-centered matrices and identical:
Thus, numerator and denominator of the non-centered CKA expression are also equal, so Gaussian CKA has the value :
| (14) |
Different intermediate numerical values occur if diagonal zeros in and are excluded from the median computation, but the final result in Eq. 14 is the same.
Now consider a linear kernel. First, compute the Gram matrices of pairwise dot products between rows:
We find non-centered linear CKA straight from the definition:
| (15) |
Eqs. 14 and 15 show that, without centering, Gaussian and linear CKA remain at a fixed positive distance from one another in this example for all bandwidths . This proves that an analog of Theorem 1 fails for non-centered CKA.
A.2 Proof of Theorem 1 in case of two Gaussian kernels
The proofs in the paper focus on the case in which one kernel is Gaussian with bandwidth and the other is a fixed kernel, . We show here how, as indicated at the end of the proof of Lemma 1, the case of two Gaussian kernels with large bandwidths follows by a triangle inequality argument. We will abbreviate as ; note that the feature representations in , are , , respectively.
First, we address Lemma 1 in the case of two Gaussian kernels. Begin by decomposing the target difference between Gaussian and Euclidean CKA as follows:
Given any desired tolerance, , the single-Gaussian case of Lemma 1 as proved in the main text (together with symmetry of CKA in its two arguments) shows that there exists a bandwidth , such that the difference term at far right, above, is less than in absolute value whenever .
Having fixed the bandwidth , there similarly exists a bandwidth such that the absolute value of the first difference term on the right-hand side above is less than whenever . By the scalar triangle inequality, it now follows that the absolute value of the target difference on the left-hand side above is smaller than whenever both and .
This argument proves Lemma 1 in the case of two Gaussian kernels: as ,
| (16) |
A.3 CKA difference for three-layer NN architecture


A.4 Results tables for section 4.2 experiments
NN width . Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| -4 | -1.28 | -1.34 | -2.25 | -0.83 | -2.21 | -0.80 |
| -3 | -0.54 | -2.04 | -3.03 | -1.75 | -2.54 | -0.40 |
| -2 | -1.29 | -2.12 | -4.08 | -3.66 | -3.58 | -1.51 |
| -1 | -3.99 | -3.12 | -5.46 | -2.92 | -4.85 | -4.64 |
| 0 | -5.52 | -4.29 | -6.70 | -3.70 | -6.87 | -6.21 |
| 1 | -5.78 | -5.41 | -8.33 | -4.97 | -8.81 | -6.84 |
| 2 | -7.30 | -6.88 | -8.12 | -6.70 | -11.00 | -8.50 |
| 3 | -9.19 | -8.70 | -8.96 | -8.63 | -12.83 | -10.42 |
| 4 | -11.17 | -10.66 | -9.97 | -10.61 | -14.82 | -12.39 |
| 5 | -13.16 | -12.64 | -11.70 | -12.61 | -16.83 | -14.39 |
| 6 | -15.16 | -14.64 | -13.64 | -14.61 | -18.84 | -16.39 |
| 7 | -17.16 | -16.64 | -15.63 | -16.61 | -20.84 | -18.39 |
| 8 | -19.16 | -18.64 | -17.62 | -18.61 | -22.84 | -20.39 |
NN width . Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| -4 | -2.29 | -0.94 | -3.08 | -2.27 | -2.73 | -2.77 |
| -3 | -1.60 | -2.18 | -4.05 | -1.28 | -3.35 | -0.60 |
| -2 | -0.99 | -2.74 | -5.23 | -4.98 | -4.62 | -0.89 |
| -1 | -3.83 | -4.04 | -6.72 | -3.87 | -6.39 | -4.01 |
| 0 | -6.39 | -5.88 | -8.43 | -4.80 | -8.30 | -7.12 |
| 1 | -6.78 | -6.68 | -10.19 | -6.37 | -10.99 | -9.20 |
| 2 | -8.30 | -8.17 | -10.01 | -8.24 | -13.46 | -10.80 |
| 3 | -10.35 | -10.05 | -10.20 | -10.20 | -15.15 | -12.75 |
| 4 | -12.19 | -12.03 | -11.45 | -12.19 | -17.1 | -14.71 |
| 5 | -14.18 | -14.02 | -13.27 | -14.19 | -19.1 | -16.69 |
| 6 | -16.17 | -16.02 | -15.22 | -16.19 | -21.11 | -18.69 |
| 7 | -18.17 | -18.02 | -17.21 | -18.19 | -23.11 | -20.69 |
| 8 | -20.17 | -20.02 | -19.21 | -20.19 | -25.11 | -22.69 |
NN width . Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| -4 | -1.57 | -0.99 | -4.30 | -3.18 | -3.21 | -3.43 |
| -3 | -3.44 | -1.18 | -5.4 | -1.78 | -4.15 | -1.32 |
| -2 | -0.86 | -4.28 | -6.69 | -4.70 | -6.03 | -0.53 |
| -1 | -3.44 | -5.30 | -7.89 | -4.81 | -8.24 | -3.42 |
| 0 | -7.45 | -5.12 | -9.74 | -5.67 | -10.49 | -6.29 |
| 1 | -8.09 | -6.32 | -11.90 | -7.25 | -13.27 | -9.53 |
| 2 | -9.34 | -8.12 | -11.71 | -9.14 | -15.92 | -11.70 |
| 3 | -11.19 | -10.06 | -11.51 | -11.12 | -18.06 | -13.91 |
| 4 | -13.15 | -12.05 | -12.87 | -13.11 | -20.29 | -15.93 |
| 5 | -15.14 | -14.05 | -14.71 | -15.11 | -22.31 | -17.96 |
| 6 | -17.14 | -16.05 | -16.67 | -17.11 | -24.24 | -19.96 |
| 7 | -19.14 | -18.05 | -18.66 | -19.11 | -26.23 | -21.97 |
| 8 | -21.14 | -20.05 | -20.66 | -21.11 | -28.23 | -23.97 |
NN width . Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| -4 | -1.53 | -0.97 | -5.84 | -2.96 | -3.82 | -2.76 |
| -3 | -1.75 | -1.04 | -7.05 | -3.04 | -5.25 | -3.80 |
| -2 | -1.03 | -2.83 | -8.33 | -5.01 | -7.65 | -0.46 |
| -1 | -3.31 | -4.17 | -9.70 | -5.24 | -10.18 | -3.13 |
| 0 | -7.09 | -4.49 | -11.86 | -6.08 | -12.56 | -5.65 |
| 1 | -9.31 | -5.94 | -13.17 | -7.72 | -15.33 | -8.56 |
| 2 | -10.32 | -7.80 | -13.25 | -9.63 | -18.46 | -11.12 |
| 3 | -12.10 | -9.76 | -13.76 | -11.61 | -20.20 | -13.27 |
| 4 | -14.05 | -11.75 | -15.07 | -13.60 | -22.23 | -15.45 |
| 5 | -16.04 | -13.75 | -16.90 | -15.60 | -24.26 | -17.33 |
| 6 | -18.04 | -15.75 | -18.86 | -17.60 | -26.27 | -19.32 |
| 7 | -20.04 | -17.75 | -20.85 | -19.60 | -28.27 | -21.32 |
| 8 | -22.04 | -19.75 | -22.85 | -21.60 | -30.27 | -23.32 |
NN width . Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| -4 | -1.43 | -0.94 | -6.75 | -3.12 | -4.30 | -2.52 |
| -3 | -1.48 | -0.97 | -7.98 | -4.65 | -5.93 | -3.99 |
| -2 | -1.31 | -2.14 | -9.24 | -5.54 | -8.42 | -0.47 |
| -1 | -3.22 | -3.25 | -10.63 | -5.35 | -10.98 | -2.88 |
| 0 | -6.27 | -4.05 | -12.57 | -6.28 | -13.40 | -5.30 |
| 1 | -10.21 | -5.59 | -14.61 | -7.94 | -16.01 | -7.96 |
| 2 | -12.02 | -7.46 | -14.16 | -9.85 | -18.90 | -10.36 |
| 3 | -13.77 | -9.43 | -14.44 | -11.83 | -21.26 | -12.50 |
| 4 | -15.70 | -11.42 | -15.80 | -13.82 | -23.34 | -14.52 |
| 5 | -17.69 | -13.42 | -17.64 | -15.82 | -25.34 | -16.52 |
| 6 | -19.67 | -15.42 | -19.61 | -17.82 | -27.34 | -18.52 |
| 7 | -21.66 | -17.42 | -21.60 | -19.82 | -29.33 | -20.52 |
| 8 | -23.66 | -19.42 | -23.59 | -21.82 | -31.33 | -22.53 |
NN width . Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| -4 | -1.34 | -0.60 | -7.68 | -3.27 | -5.05 | -2.40 |
| -3 | -1.39 | -0.63 | -8.90 | -5.91 | -7.02 | -3.27 |
| -2 | -1.16 | -1.71 | -10.17 | -5.70 | -9.59 | -0.45 |
| -1 | -3.01 | -2.81 | -11.52 | -5.51 | -12.10 | -2.68 |
| 0 | -5.71 | -3.69 | -13.38 | -6.48 | -14.47 | -5.09 |
| 1 | -9.51 | -5.23 | -14.60 | -8.15 | -17.21 | -7.63 |
| 2 | -12.15 | -7.10 | -14.53 | -10.06 | -19.76 | -9.91 |
| 3 | -14.51 | -9.06 | -14.86 | -12.04 | -21.92 | -12.00 |
| 4 | -16.45 | -11.05 | -16.14 | -14.04 | -24.06 | -14.02 |
| 5 | -18.45 | -13.05 | -17.98 | -16.03 | -26.11 | -16.02 |
| 6 | -20.45 | -15.05 | -19.94 | -18.03 | -28.13 | -18.03 |
| 7 | -22.45 | -17.05 | -21.93 | -20.03 | -30.13 | -20.03 |
| 8 | -24.45 | -19.05 | -23.93 | -22.03 | -32.13 | -22.03 |
NN width . Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| -4 | -1.40 | -0.67 | -7.96 | -3.29 | -5.01 | -2.44 |
| -3 | -1.57 | -0.72 | -9.40 | -4.01 | -6.95 | -4.48 |
| -2 | -0.72 | -1.97 | -10.78 | -4.81 | -9.55 | -0.38 |
| -1 | -2.73 | -3.06 | -11.84 | -5.81 | -12.18 | -2.56 |
| 0 | -5.28 | -3.93 | -13.24 | -6.60 | -14.67 | -4.98 |
| 1 | -8.57 | -5.46 | -13.84 | -8.23 | -17.52 | -7.49 |
| 2 | -11.26 | -7.32 | -13.95 | -10.13 | -20.21 | -9.75 |
| 3 | -13.32 | -9.29 | -14.43 | -12.11 | -22.60 | -11.83 |
| 4 | -15.36 | -11.28 | -15.89 | -14.10 | -24.46 | -13.85 |
| 5 | -17.37 | -13.28 | -17.73 | -16.10 | -26.47 | -15.86 |
| 6 | -19.37 | -15.28 | -19.70 | -18.10 | -28.48 | -17.86 |
| 7 | -21.37 | -17.28 | -21.69 | -20.10 | -30.48 | -19.86 |
| 8 | -23.37 | -19.28 | -23.69 | -22.10 | -32.49 | -21.86 |
Three-layer 128-32-8 NN. Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| -4 | -2.46 | 2.82 | -3.03 | -2.1 | -1.82 | -1.13 |
| -3 | -0.68 | 1.76 | -4.02 | -0.76 | -1.91 | -0.36 |
| -2 | -0.38 | 0.50 | -5.2 | -4.57 | -2.72 | -0.29 |
| -1 | -2.94 | -0.03 | -6.63 | -2.64 | -4.34 | -3.21 |
| 0 | -6.07 | -1.20 | -8.5 | -3.76 | -6.33 | -5.92 |
| 1 | -8.57 | -2.64 | -9.48 | -5.46 | -8.96 | -8.69 |
| 2 | -9.72 | -4.40 | -9.24 | -7.38 | -11.05 | -11.03 |
| 3 | -11.57 | -6.32 | -9.68 | -9.36 | -13.27 | -13.14 |
| 4 | -13.49 | -8.31 | -11.03 | -11.35 | -15.29 | -15.03 |
| 5 | -15.47 | -10.30 | -12.87 | -13.35 | -17.26 | -17.01 |
| 6 | -17.47 | -12.30 | -14.85 | -15.35 | -19.26 | -19.00 |
| 7 | -19.47 | -14.30 | -16.85 | -17.35 | -21.26 | -21.00 |
| 8 | -21.46 | -16.30 | -18.85 | -19.35 | -23.26 | -23.00 |
NN width . Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| -4 | -3.28 | -2.15 | -2.11 | -2.76 | -8.00 | -3.19 |
| -3 | -3.38 | -2.02 | -3.95 | -2.53 | -8.26 | -2.38 |
| -2 | -4.06 | -2.56 | -6.42 | -2.25 | -8.25 | -2.16 |
| -1 | -5.25 | -3.49 | -9.22 | -2.95 | -8.58 | -2.75 |
| 0 | -6.73 | -4.42 | -11.72 | -4.42 | -9.08 | -3.70 |
| 1 | -8.14 | -4.95 | -14.04 | -5.90 | -9.26 | -4.90 |
| 2 | -9.67 | -6.09 | -16.74 | -7.77 | -9.63 | -6.70 |
| 3 | -9.78 | -7.81 | -19.29 | -9.73 | -10.23 | -8.85 |
| 4 | -10.63 | -9.77 | -21.56 | -11.71 | -11.50 | -11.03 |
| 5 | -12.57 | -11.76 | -23.69 | -13.70 | -12.94 | -13.16 |
| 6 | -14.42 | -13.76 | -25.70 | -15.70 | -14.81 | -15.23 |
| 7 | -16.40 | -15.76 | -27.70 | -17.70 | -16.75 | -17.22 |
| 8 | -18.39 | -17.76 | -29.70 | -19.70 | -18.73 | -19.22 |
NN width . Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| -4 | -4.60 | -2.91 | -1.63 | -2.90 | -7.09 | -3.94 |
| -3 | -4.67 | -2.84 | -3.43 | -2.89 | -7.61 | -4.59 |
| -2 | -5.43 | -3.57 | -5.88 | -2.47 | -7.47 | -3.74 |
| -1 | -6.77 | -4.74 | -8.64 | -2.82 | -7.50 | -3.62 |
| 0 | -7.95 | -4.92 | -11.10 | -4.25 | -7.71 | -4.48 |
| 1 | -9.01 | -5.67 | -13.36 | -6.00 | -8.17 | -5.53 |
| 2 | -10.07 | -7.08 | -16.02 | -7.78 | -8.49 | -7.37 |
| 3 | -10.37 | -8.95 | -18.57 | -9.75 | -9.84 | -9.76 |
| 4 | -11.42 | -10.88 | -20.81 | -11.77 | -10.58 | -11.83 |
| 5 | -13.24 | -12.87 | -22.88 | -13.80 | -12.05 | -13.88 |
| 6 | -15.21 | -14.87 | -24.90 | -15.82 | -13.95 | -15.88 |
| 7 | -17.20 | -16.87 | -26.90 | -17.83 | -15.99 | -17.88 |
| 8 | -19.20 | -18.87 | -28.90 | -19.84 | -17.94 | -19.88 |
NN width . Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| -4 | -6.48 | -3.74 | -1.34 | -3.27 | -6.41 | -3.78 |
| -3 | -6.06 | -3.94 | -3.23 | -3.45 | -6.85 | -4.63 |
| -2 | -6.71 | -4.30 | -5.85 | -2.95 | -6.70 | -5.10 |
| -1 | -7.69 | -5.24 | -8.71 | -3.56 | -6.62 | -4.85 |
| 0 | -9.22 | -5.40 | -11.15 | -4.83 | -6.66 | -5.72 |
| 1 | -10.64 | -5.81 | -13.42 | -6.37 | -6.96 | -6.34 |
| 2 | -11.59 | -7.36 | -16.22 | -8.22 | -7.81 | -7.71 |
| 3 | -12.25 | -9.30 | -18.91 | -10.14 | -9.37 | -10.15 |
| 4 | -13.37 | -11.34 | -21.14 | -12.13 | -10.42 | -12.16 |
| 5 | -15.17 | -13.41 | -23.21 | -14.12 | -11.99 | -14.17 |
| 6 | -17.09 | -15.40 | -25.23 | -16.12 | -13.92 | -16.17 |
| 7 | -19.08 | -17.39 | -27.24 | -18.12 | -15.91 | -18.16 |
| 8 | -21.07 | -19.39 | -29.24 | -20.12 | -17.90 | -20.16 |
NN width . Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| -4 | -7.58 | -4.12 | -1.10 | -3.42 | -5.11 | -3.80 |
| -3 | -7.11 | -4.02 | -2.94 | -3.51 | -5.31 | -4.32 |
| -2 | -7.86 | -4.92 | -5.60 | -3.11 | -5.26 | -5.37 |
| -1 | -8.80 | -6.51 | -8.49 | -3.65 | -5.23 | -5.38 |
| 0 | -10.61 | -7.14 | -10.94 | -4.65 | -5.26 | -6.05 |
| 1 | -11.41 | -7.45 | -13.18 | -6.09 | -5.46 | -6.85 |
| 2 | -12.66 | -9.00 | -16.06 | -7.92 | -5.96 | -8.36 |
| 3 | -12.60 | -10.99 | -18.78 | -9.88 | -7.47 | -10.43 |
| 4 | -13.70 | -13.04 | -21.18 | -11.86 | -8.63 | -12.50 |
| 5 | -15.50 | -14.99 | -23.36 | -13.86 | -10.28 | -14.45 |
| 6 | -17.47 | -16.98 | -25.38 | -15.86 | -12.22 | -16.42 |
| 7 | -19.46 | -18.98 | -27.37 | -17.86 | -14.21 | -18.41 |
| 8 | -21.46 | -20.98 | -29.37 | -19.86 | -16.20 | -20.41 |
of maximum to median distance between features. is NN width. Classification.
| splice | tic-tac-toe | wdbc | optdigits | wine | dna | |
|---|---|---|---|---|---|---|
| 16 | 3.91 0.21 | 3.53 0.30 | 10.88 0.56 | 3.33 0.29 | 5.00 0.01 | 2.87 0.08 |
| 32 | 3.67 0.13 | 2.71 0.14 | 10.82 0.26 | 2.42 0.08 | 5.00 0.01 | 2.68 0.06 |
| 64 | 3.14 0.10 | 2.43 0.10 | 10.74 0.18 | 2.15 0.04 | 5.01 0.00 | 2.55 0.04 |
| 128 | 2.95 0.07 | 2.36 0.05 | 10.64 0.09 | 2.00 0.05 | 5.00 0.00 | 2.47 0.03 |
| 256 | 2.83 0.06 | 2.34 0.07 | 10.57 0.06 | 1.92 0.03 | 5.00 0.00 | 2.43 0.02 |
| 512 | 2.76 0.05 | 2.48 0.06 | 10.57 0.06 | 1.87 0.03 | 5.00 0.00 | 2.39 0.02 |
| 1024 | 2.74 0.05 | 2.45 0.04 | 10.62 0.06 | 1.89 0.03 | 5.01 0.00 | 2.39 0.02 |
| 128-32-8 | 3.28 0.12 | 4.23 0.33 | 10.87 0.26 | 2.34 0.08 | 5.00 0.00 | 2.55 0.08 |
NN width . Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| -4 | -8.24 | -4.48 | -0.90 | -2.70 | -4.23 | -3.26 |
| -3 | -7.87 | -4.47 | -2.61 | -2.74 | -4.35 | -3.56 |
| -2 | -8.71 | -5.97 | -5.28 | -2.45 | -4.33 | -5.20 |
| -1 | -9.64 | -7.36 | -8.22 | -2.94 | -4.31 | -6.21 |
| 0 | -11.44 | -7.40 | -10.70 | -4.17 | -4.32 | -6.63 |
| 1 | -12.89 | -7.67 | -13.00 | -5.66 | -4.44 | -7.99 |
| 2 | -13.74 | -9.13 | -15.68 | -7.51 | -4.81 | -8.36 |
| 3 | -14.18 | -10.99 | -18.37 | -9.46 | -6.17 | -10.52 |
| 4 | -15.26 | -12.96 | -20.66 | -11.45 | -7.44 | -12.68 |
| 5 | -17.03 | -14.95 | -22.75 | -13.45 | -9.08 | -14.69 |
| 6 | -18.97 | -16.95 | -24.78 | -15.45 | -11.00 | -16.69 |
| 7 | -20.95 | -18.95 | -26.79 | -17.45 | -12.98 | -18.69 |
| 8 | -22.95 | -20.95 | -28.79 | -19.45 | -14.98 | -20.69 |
NN width . Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| -4 | -8.29 | -3.78 | -0.75 | -2.35 | -3.68 | -2.91 |
| -3 | -8.12 | -3.80 | -2.10 | -2.39 | -3.81 | -3.25 |
| -2 | -9.00 | -5.10 | -4.40 | -2.20 | -3.78 | -5.28 |
| -1 | -9.90 | -7.37 | -7.15 | -2.64 | -3.76 | -6.29 |
| 0 | -11.48 | -7.91 | -9.61 | -3.78 | -3.79 | -6.54 |
| 1 | -13.07 | -8.15 | -11.85 | -5.31 | -3.91 | -7.45 |
| 2 | -13.93 | -9.65 | -14.89 | -7.17 | -4.24 | -7.95 |
| 3 | -15.26 | -11.56 | -18.07 | -9.13 | -5.56 | -9.79 |
| 4 | -16.44 | -13.51 | -20.29 | -11.12 | -6.86 | -11.83 |
| 5 | -18.24 | -15.49 | -22.44 | -13.12 | -8.35 | -13.85 |
| 6 | -20.22 | -17.49 | -24.47 | -15.12 | -10.25 | -15.86 |
| 7 | -22.23 | -19.49 | -26.52 | -17.12 | -12.22 | -17.86 |
| 8 | -24.23 | -21.49 | -28.50 | -19.12 | -14.22 | -19.86 |
NN width . Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| -4 | -6.87 | -3.35 | -0.67 | -2.18 | -3.26 | -2.72 |
| -3 | -6.86 | -3.19 | -1.71 | -2.22 | -3.39 | -3.11 |
| -2 | -7.90 | -4.01 | -3.64 | -2.05 | -3.35 | -5.58 |
| -1 | -8.70 | -6.49 | -5.97 | -2.46 | -3.33 | -5.28 |
| 0 | -10.07 | -7.61 | -8.20 | -3.61 | -3.36 | -6.10 |
| 1 | -11.85 | -7.65 | -10.35 | -5.15 | -3.48 | -7.13 |
| 2 | -13.22 | -8.89 | -13.34 | -7.01 | -3.78 | -7.77 |
| 3 | -15.36 | -10.72 | -16.72 | -8.98 | -4.91 | -9.62 |
| 4 | -17.14 | -12.68 | -19.23 | -10.97 | -6.16 | -11.53 |
| 5 | -18.91 | -14.67 | -21.48 | -12.97 | -7.69 | -13.53 |
| 6 | -20.81 | -16.66 | -23.63 | -14.97 | -9.58 | -15.53 |
| 7 | -22.80 | -18.66 | -25.64 | -16.97 | -11.55 | -17.53 |
| 8 | -24.80 | -20.66 | -27.63 | -18.97 | -13.55 | -19.53 |
Three-layer 128-32-8 NN. Regression.
| cpu | boston | Diab(skl) | stock | balloon | cloud | |
|---|---|---|---|---|---|---|
| -4 | -4.25 | -2.12 | -0.78 | -1.29 | -3.81 | -3.40 |
| -3 | -4.20 | -1.87 | -2.17 | -0.79 | -4.05 | -2.85 |
| -2 | -4.91 | -2.27 | -4.50 | -0.31 | -4.06 | -2.29 |
| -1 | -5.96 | -3.41 | -7.20 | -0.67 | -3.99 | -2.75 |
| 0 | -7.90 | -4.93 | -9.61 | -2.32 | -4.10 | -4.03 |
| 1 | -9.61 | -5.34 | -11.94 | -4.36 | -4.36 | -5.54 |
| 2 | -10.09 | -6.66 | -14.60 | -6.31 | -5.03 | -6.30 |
| 3 | -10.43 | -8.49 | -17.68 | -8.28 | -6.84 | -8.70 |
| 4 | -11.44 | -10.43 | -19.71 | -10.27 | -7.76 | -10.94 |
| 5 | -13.27 | -12.41 | -21.68 | -12.27 | -9.48 | -12.94 |
| 6 | -15.25 | -14.41 | -23.69 | -14.27 | -11.16 | -14.90 |
| 7 | -17.25 | -16.41 | -25.69 | -16.27 | -13.06 | -16.90 |
| 8 | -19.25 | -18.41 | -27.69 | -18.27 | -15.04 | -18.90 |