Dynamics of transposable elements generates
structure and symmetries in genetic sequences
Abstract
Genetic sequences are known to possess non-trivial composition together with symmetries in the frequencies of their components. Recently, it has been shown that symmetry and structure are hierarchically intertwined in DNA, suggesting a common origin for both features. However, the mechanism leading to this relationship is unknown. Here we investigate a biologically motivated dynamics for the evolution of genetic sequences. We show that a metastable (long-lived) regime emerges in which sequences have symmetry and structure interlaced in a way that matches that of extant genomes.
Introduction.
Transposable elements (TEs) are DNA sequences that can relocate themselves in new sites of the genome. They were firstly discovered in maize by B. McClintock in the mid-1940s and initially considered as parasites with no functional roles MC50 . Nowadays TEs are known to be ubiquitous in both prokaryotes and eukaryotes genomes FP07 ; K81 and little doubts are left of their prominent role in genome evolution, shaping structure and function in a multitude of ways BBGGetal18 ; F12 . As TEs constitute more than half of the sequence in many higher eukaryotes, a fingerprint of their presence can be quantitatively extracted from the statistical properties of their host DNA. Indeed, TEs properties were shown to be crucial in explaining structural global features of genome sequences HGBSH ; SRMA16 ; MA13 ; MAL05 ; HJ96 ; BGHPSS93 .
Recently, Albrecht-Buehler B06 suggested that TEs were the main driving force for the emergence of the second Chargaff parity rule. This rule states that, in each strand of the DNA, the frequencies of a short oligonucleotide is approximately equal to that of its symmetrically related , obtained from by reversing the order of the symbols and substituting each nucleotide with its conjugated and (e.g. , ). It has been first observed by Chargaff in the 1950s RKC68 and since then detected across different organisms leading to different proposals for its origin and function R91 ; MB06 ; NA06 ; QC01 ; FTW92 ; P93 ; BF99 ; PHB02 ; KFCHZZL09 ; ABGRPF13 ; BF99b ; LL99 ; ZH10 ; HMO12 ; CBHDK19 ; FTPM19 . The importance of Albrecht-Buehler explanation is that it shows how this symmetry naturally emerges as an asymptotic outcome of the cumulative action of inversions/transpositions, one of the main mechanism of relocation of TEs. As we will show, while the proposed mechanism nicely induces Chargaff symmetry in the asymptotic DNA, it does it at the cost of trivialisation of the structural properties of the sequence: symmetry is obtained because of the complete randomization of the full double-stranded DNA.
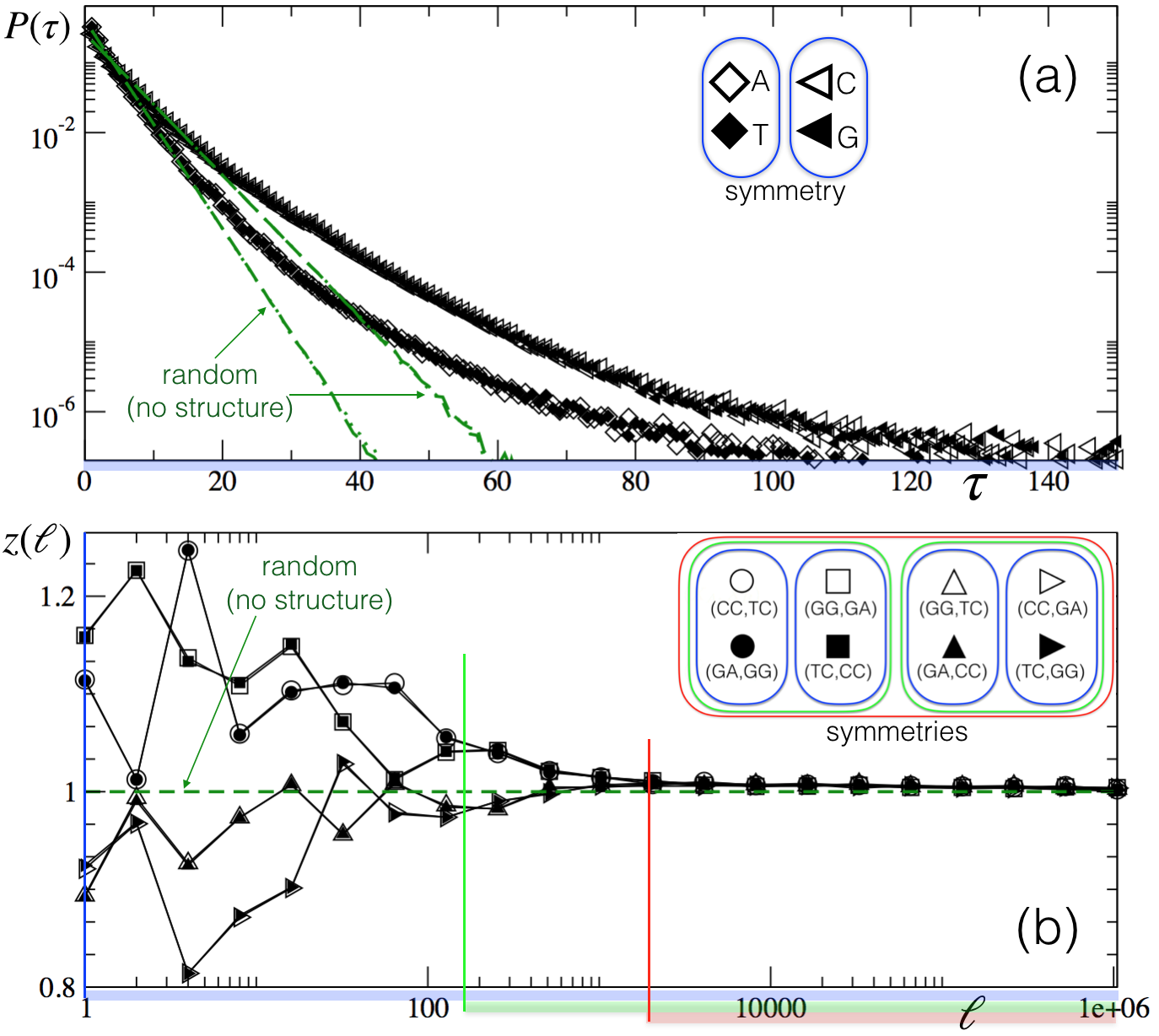
In view of the ubiquity of complex structures in genomes PBGHSSS92 ; LK92 ; V92 ; A92 ; P92 ; LMK94 ; ATVAMA01 ; FS12 ; CPTC15 , this result raises the question whether symmetry can appear without a full randomization of the sequence and in a way that is compatible with the existence of structure. The importance of this question is enhanced by our recent findings CDEA18 that Chargaff symmetry extends beyond the frequencies of short oligonucleotides – remaining valid on scales where non-trivial structure is present – and that an hierarchy of other symmetries exists, nested at different structural scales. This findings are confirmed in Fig. 1, which shows how commonly used indicators of structures, such as recurrence-time distribution (panel a) and correlation functions (panel b), coincide for symmetrically related observables at different scales.
In this work we present a biologically motivated dynamical process that explains the observed relation between symmetry and structure in DNA sequences. In particular, we propose a model that mimics the action (inversion/transpositions) of TEs on DNA and we analytically describe its dynamical behavior. Using indicators to quantify both symmetry and the presence of non-trivial structure in symbolic sequences, we show that the co-occurrence of symmetry and structure is an emergent statistical property in sequences generated by such model, reproducing the same hierarchical relation detected in extant genomes.
Quantifying structure and symmetry.
We consider symbolic sequences of length with . Given a subsequence of (a word) we denote its corresponding reverse-complemented word as , obtained from by reversing the order of the symbols and substituting each nucleotide by its complementary one and . We call the percentage of the nucleotide in the sequence . Finally, we denote by (the so called CG-content). In the following, it will be useful to partition the full set into disjoint subsets of fixed CG-content ; .
We introduce the following simple indicators of the presence of Chargaff Symmetry and of non-trivial structure composition of a given sequence .
To quantify the compliance of with Chargaff symmetry, we average the normalized difference of the abundance between a nucleotide and its symmetric one (see PHB02 where a similar measure was firstly introduced)
| (1) |
indicates a fully Chargaff-symmetric sequence, is obtained for a sequence for which Chargaff is perfectly violated (), and is obtained for a variation of equal frequencies (e.g., ). For simplicity, we consider to be a violation of Chargaff symmetry.
To quantify the presence of non-trivial structures in a given symbolic sequence we first compute the distribution of distances between two successive occurrence of the same nucleotide . For random sequences, decays exponentially as and thus has average and standard deviation (which is for small ). In contrast, the presence of a fat tail (standard deviation much larger than the mean) is considered a signature of a complex organization. We thus quantify structure as the distance of from random sequences by
| (2) |
where and are the mean and standard deviation of the measured , and is the expected for nucleotide in a random sequence. For random sequence we thus have , while departure from this value mark the presence of non-trivial structure. For simplicity, we consider to be a signature of structure.
Dynamics.
We investigate symmetry and structure of sequences that evolve through the following dynamics, that maps one sequence into another sequence by mimicking the action of TEs B06 . The dynamics is defined composing two actions:
-
(i)
pick a random position of and a random size , with 111More precisely, the pairs are drawn, independently from previous iterations, from a joint distribution chosen such that its marginal has support contained in , finite average , and the conditional distribution of positions is uniform in . We consider distributions that guarantee ergodicity of the Markov Chain. We expect ergodicity to be generically valid; e.g., it suffices to have non-zero probability for the identity transformation (i.e. ) and for single-nucleotide complementing ().
-
(ii)
replace the subsequence of size starting at position , by its reverse complement .
The couple parametrizes the effect of an inversion/transposition, which we denote by . Its action has interesting properties: is an involution for every and the total number of and (or, equivalently, of and ) is invariant under : . This implies that the dynamics is restricted to the invariant subspace of sequences with constant CG-content .
Asymptotic equilibrium.
The dynamics can be equivalently described as an ergodic Markov chain over the space of sequences . The fact that is an involution forces the transition matrix to be bi-stochastic and thus in the asymptotic equilibrium all sequences are equiprobable. This means that, for and irrespective of the initial ancient DNA sequence, the evolution asymptotically leads to sequences that can be equivalently considered generated by an independent and identically distributed (iid) process with and . Therefore, the expected value of our indicators of symmetry and structure Eqs. (1) and (2) vanish asymptotically
for any initial sequence 222We can intuitively understand this result by noting that each action of the transposon effectively creates two cuts in the sequence and moves them by a distance on average. Since cuts can happen at any location, this process eventually mixes complementary basis at different positions and breaks any correlations originally present in .. This shows analytically that the TE dynamics asymptotically leads to Chargaff symmetric sequences, in agreement with previous claims B06 . However, this symmetric equilibrium is a (trivial) consequence of a full randomization. Therefore our results show also that the current explanations of the second Chargaff parity rule B06 is not satisfactory as it is not compatible with any structure, which is known to remain significant at distances of several thousands of nucleotides PBGHSSS92 ; LK92 ; V92 ; A92 ; P92 ; LMK94 ; ATVAMA01 ; FS12 ; CPTC15 (see also Fig.1). Next we show that the same TE dynamics is rich enough by showing that symmetric sequences with non-trivial structure are generated pre-asymptotically as long-lived metastable states of TEs dynamics.
Symmetry and structure over time - three regimes.
We now investigate symmetry and structure of the sequences by computing how our indicators an depend on time (i.e., their values after applications of ). We show that Chargaff symmetry emerges much before equilibrium, together with a complex domain-like structure.
We first investigate structural properties of sequences after a finite number of iterations. We define a domain of as a subsequence of consecutive sites that have been involved in the same series of reverse/complement events. We then distinguish between domains of type and , depending on whether the number of transformations they were involved is even or odd, respectively. By definition, the starting sequence is composed by a single domain of type . After one iteration it is split into three domains, two of type and one of type of length , corresponding to the subsequence involved in the first reverse/complement event. We now compute the average sizes and of domains after iterations. Three regimes can be identified:
(i) For short times , if , the probability that the first few iterates all involve different subsequence is very high333Quantitatively, if we drop points uniformly at random on an interval of length , they will be separated by a distance at least with probability . At each iterate, a subsequence of a domain of type of average size is created, cutting a domain of type . Thus we have that in this regime:
| (3) |
This regime lasts until iterates start overlapping, which happens when and average domain-sizes equalize . This regime is thus valid for .
(ii) For a typical reverse/complement event will overlap with more than one domain. In this case all the domains that lie fully inside the subsequence involved in the reverse/complement event will change type (and position) without changing length; the domains at the border are instead split in two sub-domains of different type. The randomness of this process guarantees that the already reached balance between the number and average length of the two domains types and is not broken while their common average length decreases in time as
| (4) |
This second regime ends after a number of iterations when equilibrium is reached.
(iii) For the average lengths stabilize at the stationary value
| (5) |
and the sequence can be thought as a realization of the asymptotic equilibrium discussed above.

We now explain how structure and symmetry depend on the domain sizes and and thus on the different regimes.
-
: in order to identify the contribution of the dynamics in generating complex structural features, we consider an initial generated by an iid process (no structure, ). With this choice, a value signal the construction, under the action of the dynamics, of different domain-types. In particular, at and for , the total variance can be estimated, using the law of total variance, as the sum of two components: one that measure variability of the mean of returns between domain-types and the other measuring variability of returns within each type. Accordingly grows from to the value at the end of the first regime. In the second regime the domain sizes decay and decreases to zero at equilibrium (at ). In terms of regimes we thus expect: (i) grows; (ii) decays; (iii) .
-
: each domain of type is a subsequence of the ancient sequence . If average size of such domains at time is large enough, the frequency of each nucleotide are approximately the same as their frequency in ; similarly for and . No constraints are imposed to the symmetry of the ancient genome. In particular, if the original sequence is not Chargaff symmetric then the symmetry remains broken for all as quantified by . In terms of regimes we thus expect: (i) ; (ii) ; (iii) .
Altogether, the estimations and calculations above lead to the following predictions for the presence of symmetry and structure as a function of time (regimes i-iii):
-
(i)
Structure but no symmetry . -
(ii)
Structure and symmetry . -
(iii)
Symmetry but no structure .
In Fig. 2 we confirm these predictions in a numerical simulation.
The metastable regime.
The crucial feature of the TE dynamics discussed above is that in regime (ii) both non-trivial structure and symmetry co-exists in the generated sequences. The time (measured in number of iterations) for which this regime is valid is orders of magnitude larger than that of the first regime, as the ratio corresponds to the average size of transposable elements (for example in Homo Sapiens CBCHO07 ). We thus denote such long-lived regime as metastable and we expect it to be generically observed, even though it does not correspond to the stable equilibrium of our model.
The DNA sequences in the metastable regime are characterized by a symmetric domain-like structure. Domain models have been already introduced in literature to reproduce the complex structure generically observed in extant DNAs BGHPSS93 ; N92 ; KB93 ; PBHSSG93 ; BGRO96 ; BOFZSCMR85 ; ARLR02 ; CBCHO07 . In particular if the distribution of domain sizes has a fat tail, this will lead to a long-range correlated sequence BGHPSS93 , signalled by a slow decay of . The novelty of our approach is twofold: firstly, the domain-like structure in the metastable regime is an emergent property of the TE dynamics (it is not imposed a priori); secondly, such complex structure is intertwined with symmetry, that itself is an output of the dynamics. In particular, we have shown that sequences in the metastable regime are not only Chargaff symmetric (), they reproduce the hierarchical relation between symmetry and structure that is a distinctive feature of extant genomes (see Fig. 3 ).

Different organisms.
In Fig.4 we report and computed for genomes of different families, together with the values obtained from our dynamics. It shows that symmetry and structure coexist in most cases. The sequences from Animals shows enhanced structure while the cases of Archaea and Bacteria shows a moderate signatures of structure, in agreement with the temporal behaviour of our model (i.e., associating with the age of the genomes). Note that symmetry and structure properties are both statistical observations we made on the full DNA sequence. Any evolutionary constraint that pertains a small percentage of an organism genome does not affect these statistical observation in a sensible way. As an example, the protein-coding regions of Homo-Sapiens account for of the full sequence. On the other hand, care should be taken when dealing with many different organisms: extensions of the model incorporating additional aspects of DNA evolution will be required for a quantitative comparison with the empirical data.
Conclusion.
We have shown how a model that captures the action of transposable elements (TEs) is able to reproduce the intricate relation between symmetry and structure present in DNA sequences. We find that symmetry and structure change differently at different time scales (i.e., for different number of actions of TEs). For a large (pre-asymptotic) time interval, the sequences obtained in our model show the same non-trivial structures and an hierarchy of symmetries (including Chargaff) as in actual DNA sequences (confront panels (b) of Fig.1 and Fig.3). Our mathematical model is extremely simplified and includes the essential elements to explain the onset of symmetry and structure. In particular, it mimics only a simple action of TEs (reverse-complement), ignoring the fact that TEs are classified in different families, have different properties, and act according to different mechanisms JS88 ; munoz ; JBK .

We expect that incorporating more details of the TE dynamics in our model will refine our understanding of their role in shaping statistical properties of DNA sequences, in particular in an evolutionary viewpoint that would lead to refinements in the data-model comparison presented in Fig. 4.
References
- (1) McClintock B, The origin and behavior of mutable loci in maize, Proc. Natl. Acad. Sci. USA 36 (6): 344-55 (1950).
- (2) Feschotte C, Pritham E.J., DNA Transposons and the Evolution of Eukaryotic Genomes, Annu. Rev. Genet 41: 331-368 (2007).
- (3) Kleckner N, Transposable elements in prokaryotes, Annu. Rev. Gen. 15: 341-404 (1981)
- (4) Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, Mager DL, Feschotte C, Ten things you should know about transposable elements, Genome Biology 19 (1): 199 (2018).
- (5) Fedoroff N.V., Transposable Elements, Epigenetics and Genome Evolution, Science 338, 758-767 (2012).
- (6) Holste D, Grosse I, Beirer S, Schieg P, Herzel H, Repeats and correlations in human DNA sequences Phys. Rev. E 67(6): 061913, (2003).
- (7) Sheinman, M, Ramisch, A, Massip, F, Arndt PF, Evolutionary dynamics of selfish DNA explains the abundance distribution of genomic subsequences, Scientific Reports 6 (2016).
- (8) Massip F, Arndt PF, Neutral Evolution of Duplicated DNA: An Evolutionary Stick-Breaking Process Causes Scale-Invariant Behavior, Physical Review Letters 110 (14): 148101 (2013).
- (9) Messer PW, Arndt PF, Lässig M, Solvable Sequence Evolution Models and Genomic Correlations, Physical Review Letters 94 138103 (2005)
- (10) Attard G, Hurworth A, Jack J, Language-like features in DNA: transposable element footprints in the genome EPL (Europhysics Letters) 36, 391 (1996).
- (11) Buldyrev SV , Goldberger AL, Havlin S, Peng CK, Simons M, and Stanley HE, Generalized Levy Walk Model for DNA Nucleotide Sequences, Phys. Rev. E 47, 4514-4523 (1993),
- (12) Albrecht-Buehler G, Asymptotically increasing compliance of genomes with Chargaff’s second parity rules through inversions and inverted transpositions, Proc. Natl. Acad. Sci. USA 103, 17828-17833 (2006).
- (13) Rudner R, Karkas JD, Chargaff E, Separation of B. subtilis DNA into complementary strands I. Biological properties, II. Template functions and composition as determined, III Direct analysis. Proc. Natl. Acad. Sci. USA 60, 630-635; 915-922 (1968).
- (14) Rogerson AC, There appear to be conserved constraints on the distribution of nucleotide sequences in cellular genomes. J. Mol. Evol 32, 24-30 (1991).
- (15) Mitchell D, Bridge R, A test of Chargaff’s second rule Biochem. Biophys. Res. Commun. 340, 90-94 (2006).
- (16) Nikolaou C, Almirantis Y, Deviations from Chargaff’s second parity rule in organellar DNA Insights into the evolution of organellar genomes, Gene 381, 34-41 (2006).
- (17) Qi D, Cuticchia AJ, Compositional symmetries in complete genomes, Bioinformatics 17, 557-559 (2001).
- (18) Fickett JW , Torney DC, Wolf DR Base compositional structure of genomes. Genomics 13, 1056-1064 (1992).
- (19) Prabhu VV, Symmetry observations in long nucleotide sequences, Nucleic Acids Res. 21, 2797-2800 (1993).
- (20) Bell SJ, Forsdyke DR, Accounting units in DNA. J. Theor. Biol. 197, 51-61 (1999).
- (21) Baisnée PF, Hampson S, Baldi P, Why are complementary DNA strands symmetric?, Bioinformatics 18, 1021-1033 (2002).
- (22) Kong S-G, Fan W-L, Chen H-D, Hsu Z-T, Zhou N, Zheng Bo, Lee H-C, Inverse Symmetry in Complete Genomes and Whole-Genome Inverse Duplication, PLOS one 4, e7553 (2009).
- (23) Afreixo V1, Bastos CA, Garcia SP, Rodrigues JM, Pinho AJ, Ferreira PJ, The breakdown of the word symmetry in the human genome, J. Theor. Biol. 335, 153-1599 (2013).
- (24) Bell SJ, Forsdyke DR, Deviations from Chargaff’s Second Parity Rule Correlate with Direction of Transcription, J. Theor. Biol. 197, 63-76 (1999).
- (25) Lobry JR, Lobry C, Evolution of DNA base composition under no-strand-bias condition when the substitution rates are not constant, Mol. Biol. Evol. 16, 719-723 (1999).
- (26) Zhang SH, Huang YZ, Limited contribution of stem-loop potential to symmetry of single-stranded genomic DNA, Bioinformatics 26, 478-485 (2010).
- (27) Hart A, Martínez S, Olmos FA, Gibbs Approach to Chargaff’s Second Parity Rule, Journal of Statistical Physics 146, 408-422 (2012).
- (28) Coons LA, Burkholder AB, Hewitt SC, McDonnell DP, Korach KS, Decoding the Inversion Symmetry Underlying Transcription Factor DNA-Binding Specificity and Functionality in the Genome, iScience 15, 552-591 (2019)
- (29) Fariselli P, Taccioli C, Pagani L, Maritan A, DNA sequence symmetries from randomness: the origin of the Chargaff’s second parity rule, Briefings in Bioinformatics, bbaa041, (2020).
- (30) Peng CK , Buldyrev SV,Goldberger AL , Havlin S, Sciortino F, Simons M and Stanley HE, Long-range correlation in nucleotide sequences, Nature 356, 168-170 (1992).
- (31) Li W, Kaneko K, Long-Range Correlation and Partial Spectrum in a Noncoding DNA Sequence, EPL 17, 655-660 (1992).
- (32) Voss R, Evolution of Long-Range Fractal Correlations and Noise in DNA Base Sequences, Phys. Rev. Lett. 68, 3805-3808 (1992).
- (33) Amato I, DNA shows unexplained patterns writ large, Science 257, 747(1992).
- (34) Yam P, Noisy nucleotides: DNA sequences show fractal correlations, Sci. Am. 267, 23-24,27 (1992).
- (35) Li W, Marr TG, Kaneko K, Understanding long-range correlations in DNA sequences, Physica D 75, 392-416 (1994).
- (36) Audit B, Thermes C, Vaillant C, d’Aubenton-Carafa J, Muzy JF and Arneodo A, Long-Range Correlations in Genomic DNA: A Signature of the Nucleosomal Structure, Phys. Rev. Lett. 86, 2471 (2001).
- (37) Frahm KM, Shepelyansky DL, Poincaré recurrences of DNA sequences, Phys. Rev. E 85, 016214 (2012).
- (38) Colliva, A, Pellegrini R, Testori A, Caselle M, Ising-model description of long-range correlations in DNA sequences, Phys. Rev. E 91: 052703 (2015).
- (39) Cristadoro G, Degli Esposti M, Altmann EG, The common origin of symmetry and structure in genetic sequences, Scientific Reports 8(1), 15817 (2018).
- (40) Nee S, Uncorrelated DNA walks, Nature 357, 450 (1992).
- (41) Karlin S, Brendel V, Patchiness and correlations in DNA sequences, Science 259, 677-680 (1993).
- (42) Peng CK, Buldyrev SV, Havlin S, Simons M, Stanley HE and Goldberger AL, Mosaic organization of DNA nucleotides, Phys. Rev. E 49, 1685-1689 (1994).
- (43) Bernaola-Galván P, Román-Roldán R, Oliver JL, Compositional segmentation and long-range fractal correlations in DNA sequences, Phys. Rev. E 53, 5181-5189 (1996).
- (44) Bernardi G, Olofsson B, Filipski J, Zerial M, Salinas J, Cuny G, Meunier-Rotival M and Rodier F, The mosaic genome of warm-blooded vertebrates, Science 228, 953-958 (1985).
- (45) Rajeev K, Azad J, Subba R, Wentian Li, and Ramakrishna R, Simplifying the mosaic description of DNA sequences, Phys. Rev. E 66, 031913 (2002).
- (46) Carpena P , Bernaola-Galván P, Coronado AV, Hackenberg M, Oliver JL, Identifying characteristic scales in the human genome, Phys. Rev. E 75, 032903 (2007).
- (47) Jurka J, Smith T, A fundamental division in the Alu family of repeated sequences, Proc Natl Acad Sci U S A 85, 4775–8 (1988).
- (48) Muñoz-López M, García-Pérez JL, DNA transposons: nature and applications in genomics, Current genomics 11(2), 115–128 (2010).
- (49) Jurka J, Bao W, Kojima KK, Families of transposable elements, population structure and the origin of species, Biology direct 6, 44 (2011).
- (50) All sequences were downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). The sequences were processed to remove all letters different from A, C, G, T. The first 100000 subsequence of each entry was used for data in Fig.4.