Diffusion Methods for Generating Transition Paths ††thanks: This work is supported in part by National Science Foundation via award NSF DMS-2012286 and DMS-2309378.
Abstract
In this work, we seek to simulate rare transitions between metastable states using score-based generative models. An efficient method for generating high-quality transition paths is valuable for the study of molecular systems since data is often difficult to obtain. We develop two novel methods for path generation in this paper: a chain-based approach and a midpoint-based approach. The first biases the original dynamics to facilitate transitions, while the second mirrors splitting techniques and breaks down the original transition into smaller transitions. Numerical results of generated transition paths for the Müller potential and for Alanine dipeptide demonstrate the effectiveness of these approaches in both the data-rich and data-scarce regimes.
1 Introduction
A challenge arises in the study of molecular dynamics when the behavior of a system is characterized by rare transitions between metastable states. Practically, these rare transitions mean that Monte Carlo simulations take a prohibitively long time even if enhanced sampling techniques are used.
A common way to understand the transition process is to sample transition paths from one metastable state to the other and use this data to estimate macroscopic properties of interest. Sampling through direct simulation is inefficient due to the high energy barrier, which has led to the exploration of alternative methods. Some notable methods include transition path sampling [BCDG02], biased sampling approaches [PS10], and milestoning [FE04]. Across different applications of rare event simulation, importance sampling and splitting methods are notable. The former biases the dynamics to reduce the variance of the sampling [Fle77], while the latter splits rare transitions into a series of higher probability steps [CGR19].
Broadly, generative models such as Generative Adversarial Models (GANs) and Variational Autoencoders (VAEs) have been developed to learn the underlying distribution of a dataset and can generate new samples that resemble the training data, with impressive performance. Among other tasks, generative models have been successfully used for image, text, and audio generation. Recently, VAEs have been used for transition path generation [LRS+23]. For this approach, the network learns a map from the transition path space to a smaller latent space (encoder) and the inverse map from the latent space back to the original space (decoder). The latent space is easier to sample from and can be used to generate low-cost samples by feeding latent space samples through the decoder.
Another promising method from machine learning frequently used in image-based applications is diffusion models or score-based generative models [SSDK+20] [CHIS23]. This paper will provide methods to generate transition paths under overdamped Langevin dynamics using score-based generative modeling. As with VAEs, diffusion models rely on a pair of forward and backward processes. The forward process of a diffusion model maps the initial data points to an easy-to-sample distribution through a series of noise-adding steps. The challenge is to recover the reverse process from the noisy distribution to the original distribution, which can be achieved by using score matching [HD05] [Vin11]. Having estimated the reverse process, we can generate samples from the noisy distribution at a low cost and transform them into samples from the target distribution.
The naive application of score-based generative modeling is not effective because of the high dimensionality of discretized paths. We introduce two methods to lower the dimensionality of the problem, motivated by techniques that have been used previously for simulating transition paths. The chain method, which is introduced in Section 4.1, updates the entire path together and relies on a decomposition of the score in which each path point only depends on adjacent path points. The midpoint method, which we outline in Section 4.2, generates path points separately across multiple iterations. It also uses a decomposition which gives the probability of the midpoint of the path conditioned on its two endpoints.
In Section 2, we give an overview of diffusion models and the reverse SDE sampling algorithm for general data distributions. In Section 3, we give background on transition paths and transition path theory, before discussing the two methods for generating transition paths in Section 4. Numerical results and a more detailed description of our algorithm are included in Section 5. The main contributions of this paper are to establish that diffusion-based methods are effective for generating transition paths and to propose a new construction for decomposing the probability of a transition path for dimension reduction.
2 Reverse SDE Diffusion Model
There are three broad categories commonly used for diffusion models. Denoising Diffusion Probabilistic Models (DDPM) [HJA20] noise the data via a discrete-time Markov process with the transition kernel given by , where the hyperparameters determine the rate of noising. The reverse kernel of the process is then approximated with a neural network. The likelihood can’t be calculated, so a variational lower bound for the negative log-likelihood is used. NCSM [SE19] uses a different forward process, which adds mean-0 Gaussian noise. NCSM uses annealed Langevin sampling for sample generation, where the potential function is an approximation of the time-dependent score function . The third approach models the forward and reverse processes as an SDE [SSDK+20]. The model used in this paper can be described in either the DDPM or the reverse SDE framework, but we will describe it in the context of reverse SDEs.
Let us first review the definitions of the forward and backward processes, in addition to describing the algorithms for score-matching and sampling from the reverse SDE. Reverse SDE diffusion models are a generalization of NCSM and DDPM. As for all generative modeling, we seek to draw samples from the unknown distribution , which we have samples from. In the case of transition paths, the data is in the form of a time series. Since diffusion models are commonly used in computer vision, this will often be image data. The forward process of a reverse SDE diffusion model maps samples from to a noisy distribution using a stochastic process such that
| (1) |
where is standard Brownian motion. We denote the probability density of as . In this paper, we will use an Ornstein-Uhlenbeck forward process with . Then, and as . This means that as long as we choose a large enough , we can start the reverse process at a standard normal distribution. The corresponding reverse process is described as follows:
| (2) |
where time starts at and flows backward to , i.e., "" is a negative time differential. Reversing the direction of time we can get the equivalent expression
| (3) |
is the score function of the noised distribution, which can’t be retrieved analytically. So, the computational task shifts from approximating the posterior directly, which is the target of energy-based generative models, to approximating the noised score . Posterior estimation of complex distributions is a well-studied and challenging problem in statistics. Modeling the score is often more tractable and does not require calculating the normalizing constant.
2.1 Approximating the Score Function
In reverse SDE denoising, we use a neural network to parameterize the time-dependent score function as . Since the reverse SDE involves a time-dependent score function, the loss function is obtained by taking a time average of the distance between the true score and . Discretizing the SDE with time steps , we get the following loss function:
| (4) |
Using the step size is a natural choice for weighting, although it is possible to use different weights. The loss function can be expressed as an expectation over the joint probability of as [Vin11]
| (5) |
We can calculate based on the forward process (in the case of Ornstein-Uhlenbeck, is a Gaussian centered at ). Remarkably, this depends only on , , and the choice of forward SDE. A similar loss function is used for NCSM with a different noise-adding procedure. The training procedure follows from the above expression for the loss.
2.2 Sampling from Reverse SDE
Once we have learned the time-dependent score, we can use it to sample from the original distribution using the reverse SDE. Substituting our parameterized score function into (2), we get an approximation of the reverse process,
| (6) |
We want to evaluate at the times at which it was trained, so we discretize the SDE from equation (2) using the sequence of times , where . In forward time (flowing from 0 to T), this discretization gives
| (7) |
We solve for using Itô’s formula to retain the continuous dynamics of the first term. Let and be a standard Gaussian random variable, then
| (8) |
After solving for , we can get an explicit expression for , which is known as the exponential integrator scheme [ZC22].
| (9) |
where . We denote the distribution of as . According to the literature [CLL23], using an exponentially decaying step size for the discretization points provides strong theoretical guarantees for the distance between and the true data distribution, which we will explore further in Section 4.3. For the forward process, this means that . It is important to choose , and carefully, as they significantly affect the performance. More details about our implementation can be found in Section 5. Having estimated the score function, we can generate samples from a distribution that is close to by setting , then repeatedly applying (9) at the discretization points. A visualization of the distribution of at different values under this procedure is shown in Figure 1.

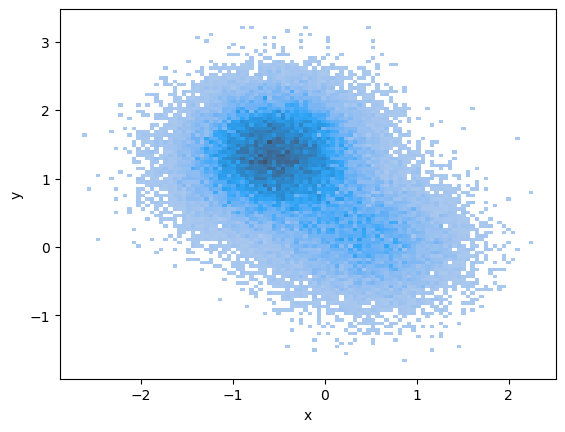


3 Transition Paths
3.1 Overdamped Langevin Dynamics
Transitions between metastable states are crucial to understanding behavior in many chemical systems. Metastable states exist in potential energy basins and so transitions will often occur on a longer time scale than the random fluctuations within the system. Due to their infrequency, it is challenging to effectively describe these transitions. In the study of transition paths, it is often useful to model the governing system as an SDE. In this section, we will look at the overdamped Langevin equation, given by
| (10) |
where is the potential of the system and is d-dimensional Brownian motion. Let be the filtration generated by . In chemical applications, is the temperature times the Boltzmann constant. For reasonable , the invariant probability distribution exists and is given by
| (11) |
We can see an example of samples generated from the invariant distribution when is the Müller potential in the above figure.
Consider two metastable states represented by closed regions . Their respective boundaries are . The paths that go from the boundary of to the boundary of without returning to are called transition paths [EVE10]. This means that a realization is a transition path if , and . The distribution of transition paths is the target distribution of the generative procedure in our paper. A similar problem involves trajectories with a fixed time interval and is known as a bridge process. Much of the analysis in this paper can be extended to the fixed time case by removing the conditioning on path time during training and generation.
3.2 Distribution of Transition Paths
We will now examine the distribution of transition paths as in [LN15]. Consider stopping times of the process with respect to :
| (13) | |||
| (14) |
Let be the event that . We are only looking at choices of such that and . It follows from Itô’s formula that is a solution to (12). Thus, the committor gives the probability that a path starting from a particular point reaches region before region .
Consider the process , the corresponding measure , and the stopping times . Since the paths we are generating terminate after reaching , we will work with the process, though it is possible to use the original process as well. We define as the event that . The function defined as is equivalent to the committor for the process. We are interested in paths drawn from the measure on transition paths. The Radon-Nikodym derivative is given by
| (15) |
Suppose that we have a path starting at and ending in . Eq. (15) states that the relative likelihood of under compared to increases as decreases. This follows our intuition, since as decreases, a higher proportion of paths starting from from the original measure will end in rather than .
4 Generating Transition Paths with Diffusion Models
We can generate transition paths by taking sections of an Euler-Maruyama simulation such that the first point is in basin A, the last point is in basin B, and all other points are in neither basin. This discretizes the definition of transition paths discussed earlier. We will represent transition paths as , where is the number of points in the path. We will denote the duration of a particular path as . This should not be confused with , which represents the duration of the diffusion process for sample generation. It is convenient to standardize the paths so that they contain the same number of points, and there is an equal time between all subsequent points in a single path.
4.1 Chain Reverse SDE Denoising
In this section, we introduce chain reverse SDE denoising, which learns the gradient for each point in the path separately by conditioning on the previous point. Specifically, we will use to represent the component of score function which corresponds to and as the corresponding neural network approximation. Unlike in the next section, here we are approximating the joint probability of the entire path (that is, ) and split it into a product of conditional distributions involving neighboring points as described in Figure 2. This is similar to the approach used in [HDBDT21], but without fixing time. Our new loss function significantly reduces the dimension of the neural network optimization problem:

| (17) |
where now denotes the th point in the path. This is a slight change in notation from the previous section, where the subscript was the time in the generation process. The time flow of generation is now represented by . We can take advantage of the Markov property of transition paths to get a simplified expression for the sub-score for interior points,
| (18) |
It follows that we need two networks, and . The first to approximate and the second for . The first and last points of the path require slightly different treatment. From the same decomposition of the joint distribution, we have that
| (19) | ||||
| (20) |
Then, the entire description of the sub-score functions is given by
| (21) |
where is a third network to learn the distribution of initial points. As outlined in Section 2.2, we use a discretized version of the reverse SDE to generate transition paths.
4.2 Midpoint Reverse SDE Denoising
While the previous approach can generate paths well, there are drawbacks. It requires learning score functions since the score for a single point is affected by its index along the path. There is a large degree of correlation for parts of the path that are close together, but it is still not an easy computational task. Additionally, all points are updated simultaneously during generation, so the samples will start in a lower data density region. This motivates using a midpoint approach, which is outlined in Figure 3.

We train a score model that depends on both endpoints to learn the distribution of the point equidistant in time to each. This corresponds to the following decomposition of the joint density. Let us say that there are points in each path. We start with the case in which the path is represented by a discrete Markov process .
Lemma 4.1.
Define , then
Proof.
First, we want to show that . We proceed by induction. The inductive hypothesis is that .
Based on this, we can parameterize the score function by
| (22) |
where . It is worth mentioning that without further knowledge of the transition path process, we would need to include as a parameter for . However, we can see from (16) that transition paths are a time-homogeneous process, which allows this simplification. Then,
| (23) | ||||
| (24) | ||||
| (25) |
where is the transition kernel for transition paths and are general functions designed to show the parameter dependence of . The training stage is similar to that described in the previous section. For an interior point , the corresponding score matching term is . For the endpoints, we have and . It is algorithmically easier to use discretization points for the paths, but we can generalize to points. Starting with the same approach as before, we will eventually end up with a term that can not be split by a midpoint if we want the correct number of points. Specifically, this occurs when we want to generate interior points for some even . In this case, we can use
| (26) |
where or . The score function will require an additional time parameter now that the midpoint is not exactly in between the endpoints. A natural choice is to use
where
| (27) |
A similar adaptation can be made when the points along the path are not evenly spaced. It remains to learn , which is a simple, one-dimensional problem. We can then use times drawn from the learned distribution as the seed for generating paths.
We can extend the result from Lemma (4.1) to the continuous case. Consider a stochastic process as in (1) and arbitrary times .
Corollary 4.2.
Define
The proof follows by the same induction as in the previous case, since the Markov property still holds. This technique can be extended to problems similar to transition path generation, such as other types of conditional trajectories. It is also possible to apply the approach of non-simultaneous generation to the chain method that we described in the previous section.
4.3 Convergence Guarantees for Score-Matching
We seek to obtain a convergence guarantee for generating paths from the reverse SDE using the approach outlined in Section 2.2. There has been previous work on convergence guarantees for general data distributions by [LLT23], [CLL23], [DB22], [CCL+22]. In particular, we can make guarantees for the KL divergence or TV distance between and the distribution of generated samples given an error bound on the score estimation. Recent results [BDBDD23] show that a bound with linear -dependence is possible for the number of discretization steps required to achieve .
Assumption 1.
The error in the score estimate at the selected discretization points is bounded:
| (28) |
Assumption 2.
The data distribution has a bounded second moment
| (29) |
With these assumptions on the accuracy of the score network and the second moment of , we can establish bounds for the KL-divergence between and . Using the exponential integrator scheme from Section 2.2, the following theorem from [BDBDD23] holds:
Theorem 4.3.
Suppose that Assumptions 1 and 2 hold. If we choose and , then there exists a choice of from 2.2 such that .
It is important to keep in mind that the KL-divergence between and does not entirely reflect the quality of the generated samples. In a practical setting, there also must be sufficient differences between the initial samples and the output. Otherwise, the model is performing the trivial task of generating data that is the same or very similar to the input data.
5 Results
5.1 Müller Potential
To exhibit the effectiveness of our algorithms, we look at the overdamped Langevin equation from (10) and choose as the two-welled Müller potential in defined by
| (30) |
We used the following parameters, as used in [KLY19] [LLR19]
| (5.2) | |||
We define regions A and B as circles with a radius of 0.1 centered around the minima at (0.62, 0.03) and (-0.56, 1.44) respectively. This creates a landscape with two major wells at A and B, a smaller minimum between them, and a potential that quickly goes to infinity outside the region . We used for this experiment, which is considered a moderate temperature.
For each of the score functions learned, we used a network architecture consisting of 6 fully connected layers with 20, 400, 400, 200, 20, and 2 neurons, excluding the input layer. The input size of the first layer was 7 (2 data points, and ) for the chain method and 8 for the midpoint method (3 data points, and ). We used a hyperbolic tangent (tanh) for the first layer and leaky ReLU for the rest of the layers. The weights were initialized using the Xavier uniform initialization method. All models were trained for 200 epochs with an initial learning rate of 0.015 and a batch size of 600. For longer paths, we used a smaller batch size and learning rate because of memory limitations. The training set was about 20,000 paths, so this example is in the data-rich regime. All noise levels were trained for every batch. We used the (9) to discretize the reverse process. We found that and with 100 discretization points gave strong empirical results. For numerical reasons, we used a maximum step size of 1. In particular, this restriction prevents the initial step from being disproportionately large, which can lead to overshooting.

The paths generated using either method (sample shown in Figure 4) closely resemble the training paths. To evaluate the generated samples numerically, we can calculate the relative entropy of the generated paths compared to the training paths or the out-of-sample paths (Tables 5a, 5b). We converted collections of paths into distributions using the scipy gaussian_kde function.
| # Discretization Points | Output and Training | Output and OOS | Training and OOS |
|---|---|---|---|
| 9 | 0.018 | 0.068 | 0.031 |
| 17 | .020 | 0.078 | 0.029 |
| 33 | 0.032 | 0.109 | 0.035 |
| # Discretization Points | Output and Training | Output and OOS | Training and OOS |
|---|---|---|---|
| 9 | 0.012 | 0.056 | 0.031 |
| 17 | 0.016 | 0.065 | 0.029 |
| 33 | 0.025 | 0.035 | 0.081 |
We can also test the quality of our samples by looking at statistics of the generated paths. For example, we can look at the distribution of points on the path. From transition path theory [LN15], we know that , where is the original probability density of the system, is the density of points along transition paths, and is the committor. This gives a 2-dimensional distribution as opposed to a dimensional one since each of the path points is treated independently. Here, the three relevant distributions are the training, output, and ground truth distributions.


We include the average absolute difference between the three relevant distributions at 2500 grid points in Tables 6a, 6b.
| # Discretization Points | Output and Training | Output and True | Training and True |
|---|---|---|---|
| 9 | 0.027 | 0.055 | 0.034 |
| 17 | 0.031 | 0.060 | 0.034 |
| 33 | 0.039 | 0.072 | 0.037 |
| # Discretization Points | Output and Training | True and Training | Output and True |
|---|---|---|---|
| 9 | 0.022 | 0.051 | 0.034 |
| 17 | 0.027 | 0.056 | 0.034 |
| 33 | 0.034 | 0.062 | 0.037 |
The chain method slightly outperforms the midpoint method according to this metric. Qualitatively, the paths generated using the two methods look similar. There is a strong correlation between the error and the training time, but a limited correlation with the size of the training dataset after reaching a certain size. This is promising for potential applications, as the data requirements are not immediately prohibitive.

The strong dependence on training time, which is shown in Figure 6, supports the notion that the numerical results of this paper could be improved with longer training or a larger model. We further explore performance in the data-scarce case in the following section.
5.2 Alanine Dipeptide
For our second numerical experiment, we generate transitions between stable conformers of Alanine dipeptide. The transition pathways [JW06] and free-energy profile [VV10] of Alanine dipeptide have been studied in the molecular dynamics literature and are shown in Figure 7. This system is commonly used to model dihedral angles in proteins. We use the dihedral angles, and (shifted for visualization purposes), as collective variables.
The two stable states that we consider are the lower density state around (minimum A), and the higher density state around (minimum B). The paths are less regular than with Müller potential because the reactive paths "jump" from values close to and . We use 22 reactive trajectories as training data for the score network. The data include 21 samples that follow reactive pathway 1 and one sample that follows reactive pathway 2. We use the same network architecture as used for the midpoint approach in the previous section. We train the model for 2000 epochs with an initial learning rate of 0.0015.
Because the data are angles, the problem moves from to the surface of a torus. We wrap around points outside of the interval during the generation process to adjust for the new topology. This approach was more effective than encoding periodicity into the neural network. For this problem, the drift term for the forward process, , is no longer continuous, though this did not cause numerical issues. It may be useful to use the more well-behaved process applied to problems with this geometry.





We can generate paths across both reactive channels. In some trials, paths are generated that follow neither channel, instead going straight from the bottom left basin to minimum A. This occurred with similar frequency to paths across RP2. It is more challenging to determine the quality of the generated paths when there is minimal initial data, but it is encouraging that the qualitative shape of the paths is similar (Figure 8). Specifically, the trajectories tend to stay around the minima for a long time, and the transition time between them is short. For this experiment, the paths generated using the midpoint method were qualitatively closer to the original distribution. In particular, disproportionately large jumps between adjacent points occurred more frequently using the chain method, though both methods were able to generate representative paths.
References
- [BCDG02] Peter G Bolhuis, David Chandler, Christoph Dellago, and Phillip L Geissler. Transition path sampling: Throwing ropes over rough mountain passes, in the dark. Annual review of physical chemistry, 53(1):291–318, 2002.
- [BDBDD23] Joe Benton, Valentin De Bortoli, Arnaud Doucet, and George Deligiannidis. Linear Convergence Bounds for Diffusion Models via Stochastic Localization. arXiv preprint arXiv:2308.03686, 2023.
- [CCL+22] Sitan Chen, Sinho Chewi, Jerry Li, Yuanzhi Li, Adil Salim, and Anru R Zhang. Sampling is as easy as learning the score: theory for diffusion models with minimal data assumptions. arXiv preprint arXiv:2209.11215, 2022.
- [CGR19] Frédéric Cérou, Arnaud Guyader, and Mathias Rousset. Adaptive multilevel splitting: Historical perspective and recent results. Chaos: An Interdisciplinary Journal of Nonlinear Science, 29(4), 2019.
- [CHIS23] Florinel-Alin Croitoru, Vlad Hondru, Radu Tudor Ionescu, and Mubarak Shah. Diffusion models in vision: A survey. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2023.
- [CLL23] Hongrui Chen, Holden Lee, and Jianfeng Lu. Improved analysis of score-based generative modeling: User-friendly bounds under minimal smoothness assumptions. In International Conference on Machine Learning, pages 4735–4763. PMLR, 2023.
- [Day92] Martin V Day. Conditional exits for small noise diffusions with characteristic boundary. The Annals of Probability, pages 1385–1419, 1992.
- [DB22] Valentin De Bortoli. Convergence of denoising diffusion models under the manifold hypothesis. arXiv preprint arXiv:2208.05314, 2022.
- [EVE10] Weinan E and Eric Vanden-Eijnden. Transition-path theory and path-finding algorithms for the study of rare events. Annual review of physical chemistry, 61:391–420, 2010.
- [FE04] Anton K Faradjian and Ron Elber. Computing time scales from reaction coordinates by milestoning. The Journal of chemical physics, 120(23):10880–10889, 2004.
- [Fle77] Wendell H Fleming. Exit probabilities and optimal stochastic control. Applied Mathematics and Optimization, 4:329–346, 1977.
- [HD05] Aapo Hyvärinen and Peter Dayan. Estimation of non-normalized statistical models by score matching. Journal of Machine Learning Research, 6(4), 2005.
- [HDBDT21] Jeremy Heng, Valentin De Bortoli, Arnaud Doucet, and James Thornton. Simulating diffusion bridges with score matching. arXiv preprint arXiv:2111.07243, 2021.
- [HJA20] Jonathan Ho, Ajay Jain, and Pieter Abbeel. Denoising diffusion probabilistic models. Advances in neural information processing systems, 33:6840–6851, 2020.
- [JW06] Hyunbum Jang and Thomas B Woolf. Multiple pathways in conformational transitions of the alanine dipeptide: an application of dynamic importance sampling. Journal of computational chemistry, 27(11):1136–1141, 2006.
- [KLY19] Yuehaw Khoo, Jianfeng Lu, and Lexing Ying. Solving for high-dimensional committor functions using artificial neural networks. Research in the Mathematical Sciences, 6:1–13, 2019.
- [LLR19] Qianxiao Li, Bo Lin, and Weiqing Ren. Computing committor functions for the study of rare events using deep learning. The Journal of Chemical Physics, 151(5), 2019.
- [LLT23] Holden Lee, Jianfeng Lu, and Yixin Tan. Convergence of score-based generative modeling for general data distributions. In International Conference on Algorithmic Learning Theory, pages 946–985. PMLR, 2023.
- [LN15] Jianfeng Lu and James Nolen. Reactive trajectories and the transition path process. Probability Theory and Related Fields, 161(1-2):195–244, 2015.
- [LRS+23] Tony Lelièvre, Geneviève Robin, Innas Sekkat, Gabriel Stoltz, and Gabriel Victorino Cardoso. Generative methods for sampling transition paths in molecular dynamics. ESAIM: Proceedings and Surveys, 73:238–256, 2023.
- [PS10] F Pinski and A Stuart. Transition paths in molecules: gradient descent in pathspace. J. Chem. Phys, 132:184104, 2010.
- [SE19] Yang Song and Stefano Ermon. Generative modeling by estimating gradients of the data distribution. Advances in neural information processing systems, 32, 2019.
- [SSDK+20] Yang Song, Jascha Sohl-Dickstein, Diederik P Kingma, Abhishek Kumar, Stefano Ermon, and Ben Poole. Score-based generative modeling through stochastic differential equations. arXiv preprint arXiv:2011.13456, 2020.
- [Vin11] Pascal Vincent. A connection between score matching and denoising autoencoders. Neural computation, 23(7):1661–1674, 2011.
- [VV10] Jiri Vymetal and Jiri Vondrasek. Metadynamics As a Tool for Mapping the Conformational and Free-Energy Space of Peptides: The Alanine Dipeptide Case Study. The Journal of Physical Chemistry B, 114(16):5632–5642, 2010.
- [ZC22] Qinsheng Zhang and Yongxin Chen. Fast sampling of diffusion models with exponential integrator. arXiv preprint arXiv:2204.13902, 2022.