2 Vector Institute, Toronto, ON M5G 1M1, Canada
11email: [email protected], {dwenlong,chenmh}@student.ubc.ca [email protected]
Debiased Noise Editing on Foundation Models for Fair Medical Image Classification
Abstract
In the era of Foundation Models’ (FMs) rising prominence in AI, our study addresses the challenge of biases in medical images while the model operates in black-box (e.g., using FM API), particularly spurious correlations between pixels and sensitive attributes. Traditional methods for bias mitigation face limitations due to the restricted access to web-hosted FMs and difficulties in addressing the underlying bias encoded within the FM API. We propose a D(ebiased) N(oise) E(diting) strategy, termed DNE, which generates DNE noise to mask such spurious correlation. DNE is capable of mitigating bias both within the FM API embedding and the images themselves. Furthermore, DNE is suitable for both white-box and black-box FM APIs, where we introduced G(reedy) (Z)eroth-O(rder) (GeZO) optimization for it when the gradient is inaccessible in black-box APIs. Our whole pipeline enables fairness-aware image editing that can be applied across various medical contexts without requiring direct model manipulation or significant computational resources. Our empirical results demonstrate the method’s effectiveness in maintaining fairness and utility across different patient groups and diseases. In the era of AI-driven medicine, this work contributes to making healthcare diagnostics more equitable, showcasing a practical solution for bias mitigation in pre-trained image FMs. Our code is provided at https://github.com/ubc-tea/DNE-foundation-model-fairness.
Keywords:
Truswothy Machine Learning Fairness Xray Classification.1 Introduction
Using pre-trained models or encoders to convert complex input data into vector representations is widely used in computer vision and natural language processing (NLP) [29, 8]. This process transforms the original data into a representative low-dimensional hidden space, preserving information for downstream tasks. With the advancement of Foundation Models (FM), services like Google MedLM [20], Voyage.ai, and ChatGPT provide data embedding services, eliminating the need for specialized hardware or extensive training. Their effectiveness is especially notable in medical fields, addressing data scarcity and hardware constraints. Clinics can enhance these models with minimal effort by fine-tuning custom classifiers on compact embedding output from FM application programming interface (APIs), which provides machine learning (ML) service as a function without users to train their own models from scratch. However, the inherent biases in the APIs’ training data and their model potentially harm marginalized groups by perpetuating gender, race, and other biases, a problem that has been exposed in the NLP field [19, 28]. The biased embedding can compromise models’ performance on minority groups [5, 9]. Therefore, it is essential to develop new solutions to attain fairness in medical image embedding from pre-trained FM API.
Studies have been done to address the fairness issues in classification problems using ML models. These methods can be categorized into three approaches: 1)Model-based strategies update or remove bias-related model parameters to mitigate bias, for example, using adversarial methods [1, 24, 12]; or prune parameters that are significant for sensitive attributes (SAs) [25]; or update model to minimize the mutual information between target and SA representations using disentanglement learning [4, 2]. However, the pretrained FM API services offer users very limited control over the model parameters [10, 11]. Thus model-based approaches are infeasible under the constraints. 2)Prediction calibration-based methods analyze the prediction probability distribution of a classifier and apply different thresholds to each subgroup to reduce the discrepancy among subgroups [6, 16, 18]. These methods need custom thresholds for various tasks, classes, and groups, which limits their generalizability. Moreover, a significant amount of validation data is needed to determine the thresholds, rendering them less efficient in medical imaging applications where data scarcity is an unavoidable problem. 3)Data-based strategies alleviate bias at the pre-training stage. Re-distribution and re-weighting methods [17, 18] address unfairness by adjusting the balance of subgroups. The effectiveness of redistributed training data is limited because it can only affect the classification head, not the pre-trained FM API embedding encoder. Recent data editing methods show strong ability to reduce disparity among subgroups by removing sensitive information from the input images [12, 23]. However, can leave the model unchanged, they fail to address the underlying bias encoded within the black-box model, e.g., FM API. Moreover, these approaches either depend on disease labels and require significant computational costs. Yao et al. introduce an image editing method independent on targeted downstream tasks via sketching [27], but this method suffers from subpar performance as not learnable.
In this work, we take two unique properties of pretrained FM API into consideration. Firstly, we aim to emphasize the need to maintain the flexibility of using FM without limiting it to a specific task while maintaining good utility, thus focusing on the learnable data editing-based method that is independent of the downstream task. We choose to edit on the image space rather than on the FM API embeddings since the former provides better control on medical image fidelity and enables interpretability on the applied editing (as shown in App. 0.B). Secondly, given the constraint against modifying parameters in the pre-trained API, we commit to not changing the FM API model parameters, even with black-box access to the API. To the best of our knowledge, no effective and universal 111We refer ‘universal’ as debiasing API embeddings for various classification tasks. method has been available to address this important, timely, but challenging research question: How to remove bias on medical images when using their embeddings from a pre-trained FM API for various classification tasks?
To answer this question, we propose the Debiased Noise Editing, DNE, where the resulted edit can be shared across all subjects to eliminate SA-related information for various disease classification tasks. Specifically, we start by using a pre-trained SA classifier, trained with the same FM API embeddings. Then, we optimize a set of learnable parameters (referred DNE noise) to be added to the images by confusing the SA classifier. Furthermore, we introduce a greedy zeroth-order optimization strategy, GeZO, when APIs restrict gradient propagation in the black-box setting. Lastly, we apply this DNE noise to input images to generate fair embeddings and achieve unbiased disease classification across various disease tasks. Extensive evaluations on disease classification tasks show our method’s effectiveness in promoting fairness while preserving utility.
2 Method
2.1 Problem Setting
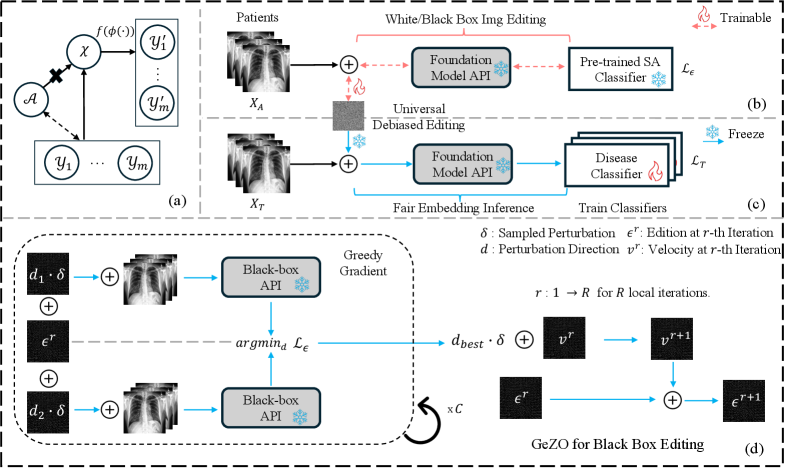
This section outlines the problem of fairness in a binary medical image classification task. Firstly, we define key variables: input images , binary disease labels , and sensitive attribute (i.e. for gender with male and female). As patients may not have sensitive and disease labels simultaneously, we denote images with sensitive attribute labels as and images with disease label as , where and are the number of samples. Then we denote the FM API as , to obtain the image embedding ; the disease classifier as and SA classifier as . We summarize all notations in App. 0.A.
Fairness Issue: Fig. 1 (a) shows there exists an association between sensitive attributes and targets (dashed line). In the Empirical Risk Minimization (ERM) [22], the model superficially consider as a proxy of , causing the issue of fairness. This phenomenon persists even when there is an equal number of data points for each class in both and , as demonstrated in Sec. 3.1. Our approach aims to address these disparities using debiased noise edit.
2.2 Debiased Noise Editing on Image for Fair Disease Classification
In this section, we present our image editing approach aimed at reducing bias via learning a noise tensor with the same dimensionality as the image, which is called DNE noise. Then, we explain how to use this DNE noise for fair disease classification.
Debiased Noise Editing. Given the spurious correlation between SA and disease , we aim to remove this link through intervention on by adding minimal editing vector , known as DNE, concealing the spurious correlations while preserving image utility. As shown in Fig. 1 (b), we first obtain a pre-trained SA classifier for medical images by either leveraging an existing one (if available) or using the collected to train the classifier with cross-entropy loss. Then, we freeze the classifier and leverage gradient ascent to learn the minimal DNE noise , which is constrained by certain threshold to preserve image fidelity. is trained to deceive the SA classifier without compromising useful information by solving the following optimization problem:
| (1) |
where regularizes the magnitude of . For example, larger enforces the small L2-norm of the in the process of gradient descent. Sec. 3.4 explores its effect in detail. Importantly, we also offer a zero-order strategy for black-box APIs, as detailed in Sec. 2.3.
Fair Disease Classification Having obtained the DNE noise to conceal the SA , we then freeze and add it to disease classification images . As shown in Fig. 1 (c), this action guarantees that the FM generates a fair embedding devoid of sensitive information, denoted as . Finally, these fair embeddings are leveraged by the subsequent disease classifier for prediction. To train the classifier , we employ a cross-entropy loss:
| (2) |
where classifier is the only trainable parameter and is the size of .
2.3 Greedy Zero-order Optimization for Black-box FM API
Zero-order (ZO) optimizations [3, 14], which avoid the need for gradient computations, offer distinct advantages in optimizing black-box models. However, these methods often exhibit slower convergence rates, particularly in training large FM models. In contrast, our proposed DNE, concentrating on the input space, offers a mitigation of this training efficiency issue of ZO optimization.
Inspired by ZO-SGD [21] and recent MeZO [15] that employ in-place perturbation updates, we propose a G(reedy) Z(ero-)O(rder) (GeZO) optimization specifically for efficient DNE. GeZO employs in-place perturbations [21, 15] and greedily updates with the gradient sign that achieves global optimal loss, thereby accelerating the optimization process. As shown in Fig. 1 (d), the core procedure of GeZO is the Greedy Gradient in the right box, where the gradient of DNE’s objective (Eq. 1) is estimated for each local iteration in GeZO. Greedy Gradient takes the DNE noise from the current local iteration () and estimates its gradient by continuously adding minor perturbations () in different directions (e.g., and ) to it, i.e., and . It then greedily selects the best direction perturbation () that results in the smallest objective in this process. Like MeZO [15], we integrate stochastic sampling for times. Each time we calculate the objective using a subset of training data to avoid sucking to the local optimum. Once the best direction and perturbation are selected for each local iteration , we add to the current velocity, . This velocity keeps track of the accumulated updating direction and magnitude for for each local iteration , where the initial value for velocity is 0. As shown in the right part of Fig. 1 (d), is updated by adding the best perturbation returned by the Greedy Gradient. Finally, the DNE noise is updated through adding the . The local iterations is a hyper-parameter, where a smaller iteration increases the efficiency at the cost of the more biased gradient estimation. We investigate the effect of different in Sec. 3.4. In actual implementation, we also introduced a momentum to update the velocity to accelerate the convergence. While the key idea is presented in this section, we provide a detailed step-by-step algorithm box for GeZO in App. 0.D.
3 Experiement
3.1 Settings
Dataset. To demonstrate the generalizability of DNE. we adopt the CheXpert dataset [7], a chest X-ray dataset with multiple disease labels, to predict the binary label for Pleural Effusion, Pneumonia and Edema individually in chest radiographs. All the ablation studies are performed on Pleural Effusion classification task for space limit. We take gender bias (male and female) as an example due to its broad impact on society and medical imaging analysis. To demonstrate the effectiveness of bias mitigation methods, we follow [4] to amplify the training data bias for each disease by (1) firstly dividing the data into different groups according to the SA; (2) secondly calculating the positive rate of each subgroup; (3) sampling subsets from the original training dataset and increase each subgroup’s bias gap (more positive sample in a subgroup). Then, we sample testing data with the same procedure, but achieve an equal subgroup bias gap. The detailed data distribution is shown in Table 3 in App. 0.C.
Evaluation metrics. We use the classification accuracy to evaluate the utility of classifiers on the test set. To measure fairness, we employ equal opportunity (EO) [6] and disparate impact (DI) [13] metrics. EO aims to ensure equitable prediction probabilities across different groups, defined by the sensitive attribute , for a given class . It quantifies the disparity in true positive rates between groups: , where a smaller gap signifies greater equality of opportunity. DI evaluates the presence of indirect discrimination by measuring the ratio of positive predictions across different groups: . DI closing to 1 indicates the minimal disparity in positive prediction rates between the groups. To align with EO metric, we employ the to quantify the fairness in percentage, with smaller values denoting greater fairness.
3.2 Implementation Details
Architecture. In our implementation, all methods use the same architecture. To simulate the FM API, we utilize a publicly available self-supervised pre-trained ViT-base model 222https://github.com/lambert-x/Medical_MAE, optimized on X-ray images, given its superior performance metrics among all other architectures [26]. Within our study, we fix the encoder of the ViT model and only fine-tune the classification head, simulating the configuration detailed in Sec. 2.1.
Debiased Noise Editing. For DNE, we first fine-tune the SA classifier, , using the feature encoded by the FM () with the training set of Pleural Effusion in Table 3. We update it using Adam with a learning rate (lr) of for epochs. Second, we update the DNE through implementing Eq. 1 by initializing as a trainable PyTorch parameter with all entries initially set to zero. The parameter matches the input image dimensions of . The DNE can be optimized using classic gradient descent or GeZO. Finally, we fine-tune the disease classifier, , with added on the input data as delineated in Eq. 2. We update it using AdamW with lr of for epochs. We provide the visualization of the magnitude of DNE as an interpretation of DNE in App. 0.B Fig. 3, where DNE is smoothed by the Gaussian kernel. As shown, larger noises are added to the bottom to discriminate the gender-related features.
GeZO. Here, we keep all parts for DNE the same as above, except using GeZO to update the edit rather than the Adam, given that the gradient is not accessible. The only key parameter that we are interested in is the local iteration , where we investigated it in Sec. 3.4. Furthermore, the detailed implementation of the algorithm and hyperparameter setting for GeZO can be found in App. 0.D.
3.3 Comparison with Baselines
| Diseases | Pleural Effusion | Pneumonia | Edema | |||||||||
| |1-DI| | Acc | |1-DI| | Acc | |1-DI| | Acc | |||||||
| ERM | 40.5 | 57.0 | 58.5 | 72.9 | 70.0 | 70.0 | 74.5 | 59.6 | 42.0 | 39.0 | 42.0 | 74.5 |
| Sketch [27] | 44.0 | 52.0 | 57.5 | 66.3 | 64.0 | 61.0 | 72.6 | 56.3 | 44.0 | 15.0 | 15.5 | 67.0 |
| Group DRO [18] | 41.0 | 58.0 | 59.5 | 72.3 | 60.0 | 56.0 | 64.4 | 61.0 | 40.5 | 40.5 | 43.3 | 74.8 |
| Batch Samp. [17] | 41.5 | 50.5 | 51.8 | 74.3 | 64.0 | 72.0 | 76.6 | 60.5 | 39.0 | 43.0 | 44.8 | 76.0 |
| BiasAdv [12] | 39.0 | 54.5 | 58.2 | 71.1 | 36.0 | 54.0 | 77.1 | 61.0 | 39.0 | 33.5 | 36.6 | 74.9 |
| DNE | 28.5 | 23.0 | 25.6 | 75.1 | 38.0 | 25.0 | 34.7 | 61.3 | 27.5 | 17.0 | 19.7 | 75.6 |
| DNE-GeZO | 37.0 | 25.0 | 28.5 | 72.8 | 35.0 | 27.0 | 33.7 | 61.5 | 34.0 | 15.5 | 17.2 | 75.1 |
3.3.1 Quantitative Analysis.
Table 1 summarizes the performances among different diseases and baselines. The baselines include model-based strategy, like biasAdv 333In BiasAdv’s implementation, we treat FM API as a white box as it requires access to FM’s gradient and there is no zeroth-order optimization for it yet. [12]; data-based strategies, like batch sampling [17] with data re-distribution and sketch [27] with data generation; and prediction calibration-based strategy, like Group DRO [18]. The details of these baselines are introduced in Sec. 1. The results indicate that the DNE effectively balances fairness and utility. Taking Pleural Effusion as the example to analyze, the white-box optimized DNE outperforms all baselines. The and DI decrease over 25% compared to all the baselines, an indication of less disparity between the two genders. Additionally, the utility not only maintains but also surpasses all other groups, e.g., DNE’s accuracy (Acc) is higher than 2.2% compared to ERM. Given we are using a balanced testing set, this increase is also a sign of better generalization. Similar for Pneumonia and Edema, where DNE and DNE-GeZO takes the best two performance for almost all entries. Although learned through black-box optimization, our DNE-GeZO achieves comparable performance to the standard optimization used in DNE. This demonstrates not only the validity of GeZO but also the efficiency afforded by the small number of edition parameters [15]. In Pneumonia, DNE-GeZO even slightly surpasses the performance of DNE across most metrics. These findings affirm the efficacy of DNE, where it not only facilitates fair medical image embedding and training but also introduces better generalizability in downstream classification tasks. Furthermore, one trained DNE can be potentially applied to all other diseases of Chest Xray, where it consistently outperforms the baselines across three diseases in both fairness and utility.
3.4 Ablation Studies
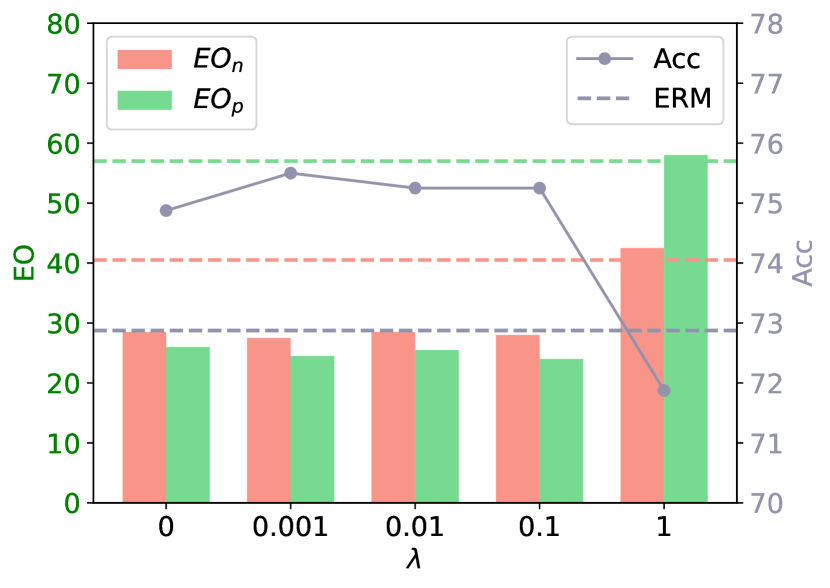
Effect of Regularization Coefficient. To investigate the effect of different regularization coefficients’ () effect, we vary from 0 to 1. Fig. 2 (a) depicts the EO and the Acc for different s. The metrics for ERM are labeled as horizontal dashed lines for convenience. As shown, both the fairness and utility metrics remain relatively stable for , consistently surpassing the ERM baseline. However, as increases to 1, we observe a marked decline in accuracy below the ERM benchmark, along with a notable increase in the EO metric for both classes.
(a)

(b)
Effect of Local Iterations in GeZO. In black-box API, total local iterations in GeZO affect the optimization performance, wherein larger leads to more accurate optimization at the cost of efficiency. Here, we examine the effect of changing , as shown in Fig. 2 (b). For fairness metrics, increasing from 2 to 20 significantly reduces the EO score for both classes, demonstrating a considerable debiasing impact with more local epochs. This effect is attributed to increased perturbation sampling that expands the search space with more local epochs, as introduced in Sec. 2.3. Meanwhile, accuracy remains relatively stable, with minor fluctuations between 72% and 73%.
4 Conclusion
In this study, we address a crucial, yet under-explored aspect of health equity—the inherent bias in FM API’s usage for classification—through the introduction of debiased noise editing. DNE effectively masks bias-inducing pixels, enhancing fairness in API-generated embeddings. Furthermore, GeZO tackles the challenge of the inaccessibility of the gradient in black-box APIs by estimating gradients via perturbation. Future research will extend DNE’s application across various FM APIs and settings, aiming to solidify its role in promoting fairer machine learning practices.
4.0.1 Acknowledgements
This work is supported in part by the Natural Sciences and Engineering Research Council of Canada (NSERC), Compute Canada, and Vector Institute.
4.0.2 \discintname
The authors have no competing interests to declare that are relevant to the content of this article.
References
- Adeli et al. [2021] Adeli, E., Zhao, Q., Pfefferbaum, A., Sullivan, E.V., Fei-Fei, L., Niebles, J.C., Pohl, K.M.: Representation learning with statistical independence to mitigate bias. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. pp. 2513–2523 (2021)
- Chen et al. [2023] Chen, M., Jiang, M., Dou, Q., Wang, Z., Li, X.: Fedsoup: improving generalization and personalization in federated learning via selective model interpolation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 318–328. Springer (2023)
- Chen et al. [2017] Chen, P.Y., Zhang, H., Sharma, Y., Yi, J., Hsieh, C.J.: Zoo: Zeroth order optimization based black-box attacks to deep neural networks without training substitute models. In: Proceedings of the 10th ACM workshop on artificial intelligence and security. pp. 15–26 (2017)
- Deng et al. [2023] Deng, W., Zhong, Y., Dou, Q., Li, X.: On fairness of medical image classification with multiple sensitive attributes via learning orthogonal representations. In: International Conference on Information Processing in Medical Imaging. pp. 158–169. Springer (2023)
- Glocker et al. [2021] Glocker, B., Jones, C., Bernhardt, M., Winzeck, S.: Algorithmic encoding of protected characteristics in image-based models for disease detection. arXiv preprint arXiv:2110.14755 (2021)
- Hardt et al. [2016] Hardt, M., Price, E., Srebro, N.: Equality of opportunity in supervised learning. Advances in neural information processing systems 29 (2016)
- Irvin et al. [2019] Irvin, J., Rajpurkar, P., Ko, M., Yu, Y., Ciurea-Ilcus, S., Chute, C., Marklund, H., Haghgoo, B., Ball, R., Shpanskaya, K., et al.: Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of the AAAI conference on artificial intelligence. vol. 33, pp. 590–597 (2019)
- Jaiswal et al. [2024] Jaiswal, A., Liu, S., Chen, T., Wang, Z., et al.: The emergence of essential sparsity in large pre-trained models: The weights that matter. Advances in Neural Information Processing Systems 36 (2024)
- Jin et al. [2024] Jin, R., Xu, Z., Zhong, Y., Yao, Q., Dou, Q., Zhou, S.K., Li, X.: Fairmedfm: Fairness benchmarking for medical imaging foundation models. arXiv preprint arXiv:2407.00983 (2024)
- Jones et al. [2024] Jones, C., Castro, D.C., De Sousa Ribeiro, F., Oktay, O., McCradden, M., Glocker, B.: A causal perspective on dataset bias in machine learning for medical imaging. Nature Machine Intelligence pp. 1–9 (2024)
- Larrazabal et al. [2020] Larrazabal, A.J., Nieto, N., Peterson, V., Milone, D.H., Ferrante, E.: Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proceedings of the National Academy of Sciences 117(23), 12592–12594 (2020)
- Lim et al. [2023] Lim, J., Kim, Y., Kim, B., Ahn, C., Shin, J., Yang, E., Han, S.: Biasadv: Bias-adversarial augmentation for model debiasing. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 3832–3841 (2023)
- Lipton et al. [2018] Lipton, Z., McAuley, J., Chouldechova, A.: Does mitigating ml’s impact disparity require treatment disparity? Advances in neural information processing systems 31 (2018)
- Liu et al. [2020] Liu, S., Chen, P.Y., Kailkhura, B., Zhang, G., Hero III, A.O., Varshney, P.K.: A primer on zeroth-order optimization in signal processing and machine learning: Principals, recent advances, and applications. IEEE Signal Processing Magazine 37(5), 43–54 (2020)
- Malladi et al. [2024] Malladi, S., Gao, T., Nichani, E., Damian, A., Lee, J.D., Chen, D., Arora, S.: Fine-tuning language models with just forward passes. Advances in Neural Information Processing Systems 36 (2024)
- Menon and Williamson [2018] Menon, A.K., Williamson, R.C.: The cost of fairness in binary classification. In: Conference on Fairness, Accountability and Transparency. pp. 107–118. PMLR (2018)
- Puyol-Antón et al. [2021] Puyol-Antón, E., Ruijsink, B., Piechnik, S.K., Neubauer, S., Petersen, S.E., Razavi, R., King, A.P.: Fairness in cardiac mr image analysis: an investigation of bias due to data imbalance in deep learning based segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part III 24. pp. 413–423. Springer (2021)
- Sagawa et al. [2019] Sagawa, S., Koh, P.W., Hashimoto, T.B., Liang, P.: Distributionally robust neural networks for group shifts: On the importance of regularization for worst-case generalization. arXiv preprint arXiv:1911.08731 (2019)
- Schramowski et al. [2022] Schramowski, P., Turan, C., Andersen, N., Rothkopf, C.A., Kersting, K.: Large pre-trained language models contain human-like biases of what is right and wrong to do. Nature Machine Intelligence 4(3), 258–268 (2022)
- Singhal et al. [2023] Singhal, K., Tu, T., Gottweis, J., Sayres, R., Wulczyn, E., Hou, L., Clark, K., Pfohl, S., Cole-Lewis, H., Neal, D., et al.: Towards expert-level medical question answering with large language models. arXiv preprint arXiv:2305.09617 (2023)
- Spall [1992] Spall, J.C.: Multivariate stochastic approximation using a simultaneous perturbation gradient approximation. IEEE transactions on automatic control 37(3), 332–341 (1992)
- Vapnik [1991] Vapnik, V.: Principles of risk minimization for learning theory. Advances in neural information processing systems 4 (1991)
- Wachinger et al. [2021] Wachinger, C., Rieckmann, A., Pölsterl, S., Initiative, A.D.N., et al.: Detect and correct bias in multi-site neuroimaging datasets. Medical Image Analysis 67, 101879 (2021)
- Wadsworth et al. [2018] Wadsworth, C., Vera, F., Piech, C.: Achieving fairness through adversarial learning: an application to recidivism prediction. arXiv preprint arXiv:1807.00199 (2018)
- Wu et al. [2022] Wu, Y., Zeng, D., Xu, X., Shi, Y., Hu, J.: Fairprune: Achieving fairness through pruning for dermatological disease diagnosis. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 743–753. Springer (2022)
- Xiao et al. [2023] Xiao, J., Bai, Y., Yuille, A., Zhou, Z.: Delving into masked autoencoders for multi-label thorax disease classification. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision. pp. 3588–3600 (2023)
- Yao et al. [2022] Yao, R., Cui, Z., Li, X., Gu, L.: Improving fairness in image classification via sketching. arXiv preprint arXiv:2211.00168 (2022)
- You et al. [2024] You, C., Min, Y., Dai, W., Sekhon, J.S., Staib, L., Duncan, J.S.: Calibrating multi-modal representations: A pursuit of group robustness without annotations. arXiv preprint arXiv:2403.07241 (2024)
- Zhou et al. [2023] Zhou, C., Li, Q., Li, C., Yu, J., Liu, Y., Wang, G., Zhang, K., Ji, C., Yan, Q., He, L., et al.: A comprehensive survey on pretrained foundation models: A history from bert to chatgpt. arXiv preprint arXiv:2302.09419 (2023)
Appendix 0.A Notation Table
| Notations | Description |
| , | input image, input space |
| , | disease label, label space |
| , | sensitive attribute label, sensitive label space |
| , | embedding, embedding Space |
| freezed FM encoder | |
| disease classifier | |
| sensitive attribute classifier | |
| universal edition | |
| regularization coefficient | |
| disease target |
Appendix 0.B Visualization

Appendix 0.C Data Distribution
| Diseases | Pneumonia | Edema | Pleural Effusion | |||||||||
| Negative | Positive | Negative | Positive | Negative | Positive | |||||||
| M | F | M | F | M | F | M | F | M | F | M | F | |
| Train Sample | 1500 | 150 | 150 | 1500 | 5000 | 500 | 500 | 5000 | 5000 | 500 | 500 | 5000 |
| Test Sample | 100 | 100 | 100 | 100 | 200 | 200 | 200 | 200 | 200 | 200 | 200 | 200 |
Appendix 0.D Greedy Zero-order Optimization
Setup: Local Iterations , step size , step size decay , Edit from last global epoch , sample times , best loss , best direction , momentum: , batch size .