Beyond Pixel-Wise Supervision for Medical Image Segmentation: From Traditional Models to Foundation Models
Abstract
Medical image segmentation plays an important role in many image-guided clinical approaches. However, existing segmentation algorithms mostly rely on the availability of fully annotated images with pixel-wise annotations for training, which can be both labor-intensive and expertise-demanding, especially in the medical imaging domain where only experts can provide reliable and accurate annotations. To alleviate this challenge, there has been a growing focus on developing segmentation methods that can train deep models with weak annotations, such as image-level, bounding boxes, scribbles, and points. The emergence of vision foundation models, notably the Segment Anything Model (SAM), has introduced innovative capabilities for segmentation tasks using weak annotations for promptable segmentation enabled by large-scale pre-training. Adopting foundation models together with traditional learning methods has increasingly gained recent interest research community and shown potential for real-world applications. In this paper, we present a comprehensive survey of recent progress on annotation-efficient learning for medical image segmentation utilizing weak annotations before and in the era of foundation models. Furthermore, we analyze and discuss several challenges of existing approaches, which we believe will provide valuable guidance for shaping the trajectory of foundational models to further advance the field of medical image segmentation.
Index Terms:
Medical Image Segmentation, Annotation-Efficient Learning, Weakly Supervised Learning, Foundation Models, Survey.I Introduction
Medical image segmentation aims to delineate the interested anatomical structures like organs and tumors from the original images by labeling each pixel into a certain class, which is one of the most representative and comprehensive research topics in the community of medical image analysis [1, 2]. Accurate segmentation can provide reliable volumetric and shape information of target structures and assist in many further clinical applications like disease diagnosis, quantitative analysis, and surgical planning [3, 4]. Since manual contour delineation is labor-intensive and time-consuming and suffers from inter-observer variability, it is highly desired in clinical studies to develop automatic medical image segmentation methods.
The advancements in deep learning have significantly leveraged the potential of deep neural networks in various medical image segmentation tasks [5, 6, 7]. Among various deep learning-based networks, U-Net [8, 9] and its variants [10, 11, 12, 13] are widely used and developed, following the encoder-decoder architecture. Specifically, nnU-Net with automatically adapted training strategies and network architectures based on U-Net architecture has shown state-of-the-art performance on many medical image segmentation tasks [14]. More recently, the success of transformer architectures in various computer vision tasks [15] spurred the development of transformer-based segmentation methods, which have been increasingly applied in the field of medical image segmentation [16, 17, 18].

Although these architectural advancements have shown encouraging results, these cutting-edge segmentation methods mainly require large amounts of training data with pixel-wise annotations [19]. However, manually annotating medical images at pixel-wise is a costly and time-consuming process, which necessitates the expertise of experienced clinical professionals. The scarcity of annotated medical imaging data is compounded by variations in patient populations, acquisition parameters, protocols, sequences, vendors, and centers, leading to significant statistical discrepancies [20]. Besides, many commonly used medical images like computed tomography (CT) and magnetic resonance imaging (MRI) are 3D volumetric data, which further increase the burden of manual annotation since experts need to delineate the target from the volume slice by slice [21]. The dense manual labeling can take several hours to annotate one image for experienced radiologists. These challenges have stimulated extensive research into developing segmentation networks with limited annotations like semi-supervised learning [22]. Despite the performance of these semi-supervised segmentation methods to utilize unlabeled data [23, 24, 25], these methods still require a subset of pixel-wise annotated data to achieve comparable performance. As an alternative, weak annotations like bounding boxes or scribbles can significantly reduce the annotation cost compared with pixel-wise fully annotations, as observed in Fig. 1. Besides, the emergence of vision foundation models [26] has also introduced innovative capabilities for segmentation tasks using weak annotations like points and bounding boxes for promptable segmentation enabled by large-scale pre-training.
In this paper, we present a comprehensive survey that encapsulates the evolution of weakly supervised medical image segmentation techniques, which have emerged as a response to the challenges associated with the high cost and expertise required for pixel-wise annotations in medical imaging. By synthesizing the findings from various studies and presenting a unified perspective on weakly supervised medical image segmentation, this paper contributes to the collective understanding of the field and offers guidance for future research endeavors.
| Reference | 2D/3D | Modality | Dataset | Strategy to bridge the supervision gap |
|---|---|---|---|---|
| Jia et al. [27] | 2D | Microscope | TMAs [28] | Incorporate multi-scale learning and area constraints for cancerous region segmentation. |
| Feng et al. [29] | 3D | CT | LIDC-IDRI [30] | Learn discriminative regions from the activation maps of convolution units at different scales. |
| Chen et al. [31] | 3D | MRI | ProMRI [32, 33], ACDC [5], CHAOS [34] | Leverage both category-causality and anatomy-causality to overcome ambiguous boundaries and co-occurrence. |
II Task Formulation
Given a defined set of classes represented by , for a given image (for 3D volumetric images) , the segmentation task aims to generate predictions , where of pixel/voxel represents the label tuple for the pixel/voxel in at the same spatial location, and and are the height, width and depth of the image, respectively, is the space of nature numbers, and represents the classification class of the pixel/voxel at spatial location .
For the training of segmentation models, given a training set with images, where represents the dense annotations of the image , which means that each pixel/voxel at spatial location of image is annotated by a label tuple . However, due to the manual labeling burden for pixel-wise annotations which is expensive and difficult to obtain, weakly supervised annotation-efficient methods focus on training segmentation models based on weak labels that cannot cover full supervision signals but are much easier to obtain. These annotation-efficient approaches are classified by the types of supervision from weak labels, which can be formulated from a unified perspective regarding the format of the training data. In the following, we introduce and give mathematical definitions for several commonly used weak annotations for medical image segmentation tasks.
Image-level Annotation. Image-level annotation is a type of annotation that utilizes only high-level image-level labels to train segmentation models. The training set can be formulated as , where is the image-level labels representing the presence or absence of each object class in the image.
Bounding-box Annotation. Bounding-box annotation aims to draw a rectangular box around the object of interest to indicate its location and size within the image. The training set can be formulated as and , where is the vertex coordinates of bounding box of the image.
Scribble/Point Annotation. Sparse annotation represents that only a discriminative subset of the target is annotated. For example, scribble annotation is a set of scribbles with only coarse-grained or incomplete annotations where only rough object boundaries are provided. Specifically, each object is represented by a set of scribbles, which are manually drawn lines along or around the object’s boundary. Point annotation aims to simply mark a few key points inside the object of interest in an image instead of drawing rough boundaries around it like in scribble annotation. The training set can be formulated as , where represents the sparse annotations including scribbles and points.
Partially Supervision. In real-world applications, a great number of datasets are partially annotated to meet different practical usages, which is commonly seen in medical image analysis. For example, most medical datasets are collected for the segmentation of only one type of organ and/or tumors, while other task-irrelevant objects are treated as the background. Partially supervised segmentation tasks have been made to utilize multiple partially labeled datasets for multi-task segmentation. The training set can be formulated as , where represents the partial annotations of class among all target classes .

| Reference | 2D/3D | Modality | Dataset | Strategy to bridge the supervision gap |
|---|---|---|---|---|
| Rajchl et al. [35] | 2D | MRI | IUGR dataset [36] | Iterative energy minimization over densely-connected conditional random field. |
| Wang et al. [37] | 3D | MRI | PROMISE12 [32], ATLAS [38] | Integrate bounding box tightness prior with smooth maximum approximation. |
| Redekop et al. [39] | 3D | CT | MSD liver [40] | Crop images within loose bounding boxes into patches for binary patch classification. |
| Tang et al. [41] | 3D | CT | NIH DeepLesion [42] | Refine regions near segmentation boundary with level-set loss. |
III Solutions for Medical Image Segmentation with Weak Annotations
III-A Medical Image Segmentation with Image-Level Annotations
Image-level annotation is one of the most efficient forms for weak-supervised learning, which provides only high-level class labels for each training image without any pixel-wise annotation. The major challenge of using image-level annotations for pixel-wise predictions lies in the significant disparity. Specifically, image-level annotations cannot provide sufficient information to guide models to accurately make pixel-wise predictions and outline interested targets.
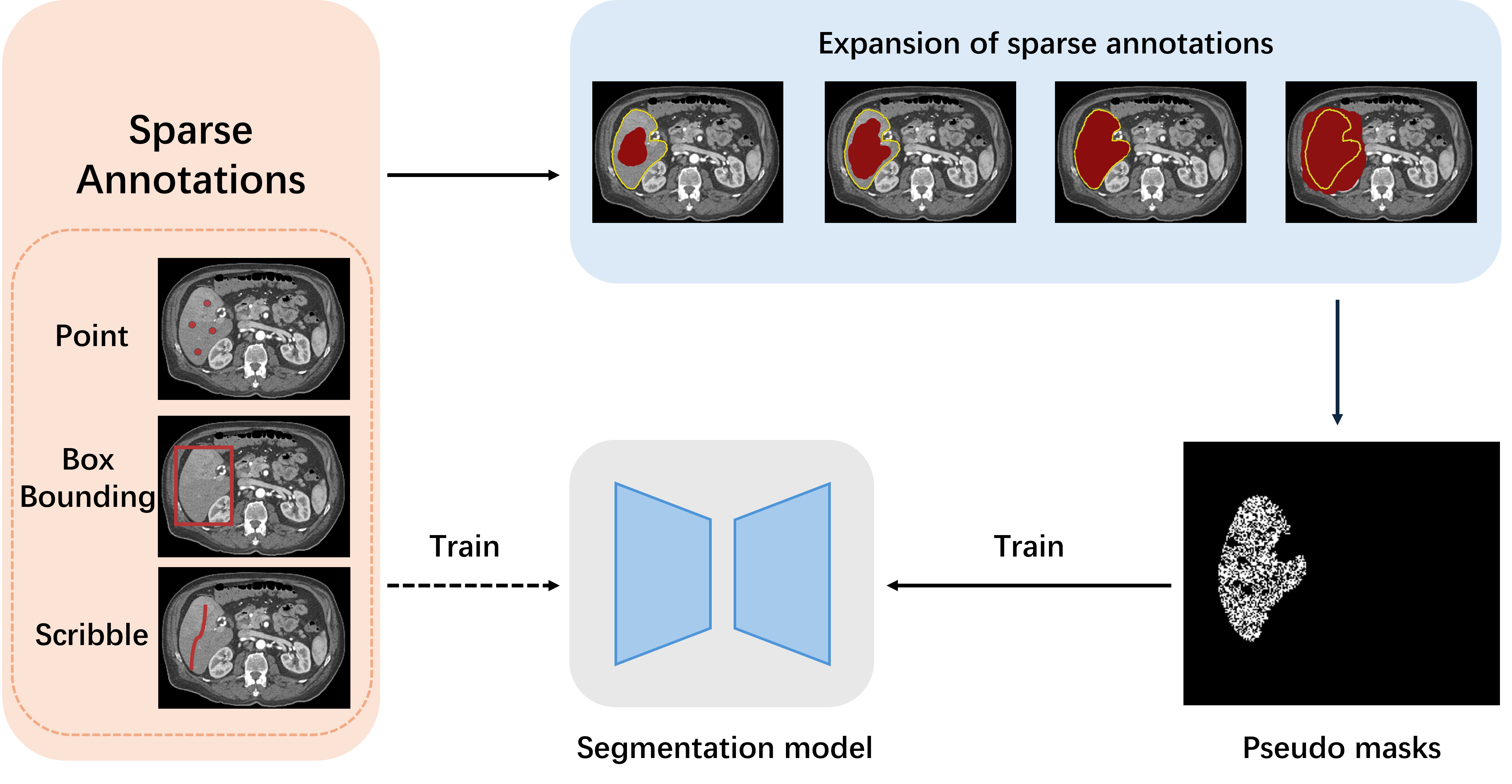
An overall workflow of weakly supervised image segmentation with image-level annotations is illustrated in Fig. 2. In the first state, pseudo masks are generated for each training image based on a classification model trained with image-level supervision. After that, a segmentation model is trained based on the pseudo masks. From the goal of applying the classification model is to generate high-quality pseudo masks from image-level annotations, which consists of two subsequent steps. Firstly, some seed areas are obtained in each training image based on the information derived from the classification model. Then the pseudo masks are generated by propagating the seed areas to the whole image. Since the pseudo masks are inevitably noisy due to the lack of supervision, the generation process is usually implemented in an iterative manner to enable the model to improve the quality of pseudo masks progressively.
For the generation of seed areas, most approaches are based on the concept of class activation mapping (CAM) [43] and its variants [44, 45, 46] achieved by classification model that covers discriminative regions of the image. Specifically, Früh et al. [47] evaluate the feasibility of weakly supervised framework by comparing different CAM methods including classic CAM [43], GradCAM [44], GradCAM++ [45] and ScoreCAM [46] in tumor segmentation with image-level annotations. Experimental results demonstrate that CAM, GradCAM++, and ScoreCAM yielded similar results, while GradCAM led to inferior results that may ignore multiple instances in a given slice. Despite serving as an efficient tool to generate seed areas based on the classification model, CAM still suffers from several limitations. For example, CAM could only locate the discriminative part of the object and could not completely cover the object, which may deteriorate the performance. Additionally, compared with natural images, medical images usually have low contrast resulting in objects have ambiguous boundaries. Therefore, directly applying CAM-based methods is usually less effective. Besides, the co-occurrence is very severe in medical images, which means that different segmentation targets (e.g. organs) often occur in the same image [31]. As a result, it is hard to activate correct co-occurring targets in one image only according to image-level labels.
To address these challenges, several approaches are proposed in refining seed areas to make them more accurate and generate more reliable pseudo masks based on the seed areas. Li et al. [48] propose a weakly-supervised framework with image-level annotations for breast tumor segmentation. Breast tumors are recognized by a classification model and then segmented based on class activation mapping and deep level set (CAM-DLS), where domain-specific anatomical information of breast ultrasound is utilized to reduce the search space. Izadyyazdanabadi et al. [49] construct a weakly supervised framework to generate glioma Diagnostic Feature Maps (DFM) from confocal laser endomicroscopy (CLE) images using a multi-layer CAM followed by global average pooling to obtain the image-level label prediction. Hwang et al. [50] present a self-transfer learning (STL) framework for weakly supervised lesion localization by jointly optimizing classification and localization networks to help the localization network focus on correct lesions without any types of priors. Yoo et al. [51] propose a pipeline to use binary classification labels to segment regions of interest (ROIs) for brain tumor segmentation using MRI scans. Feng et al. [29] propose a two-stage weakly supervised framework that involves a coarse image segmentation followed by a fine-grained instance-level segmentation. The first stage leverages CAM generated by an image classification model to determine the presence or absence of a nodule in each slice. Then, in the second stage, the region of interest around each localized instance in the class activation map is selected while everything outside this region is masked out. Jia et al. [27] propose a multiple instance learning framework for weakly supervised segmentation given only image-level cancer labels for histopathology images by introducing constraints about positive instances to effectively explore additional weakly supervised information. Each histopathology image is treated as a bag and each individual pixel in the image is treated as an instance. To regularize training, area constraints are further imposed to the segmentation results of multiple-instance learning. Dubost et al. [52] propose a new weakly supervised framework to compute attention maps that reveal the locations of brain lesions. using the last feature maps of a segmentation network optimized only with global image-level annotations. Dang et al. [53] propose a novel annotation-efficient framework to utilize weak patch-level tag labels and a CAPTCHA-like setup to synthesize pixel-wise pseudo-labels for vessel and background. Chen et al. [31] propose a causal class activation map (C-CAM) method for weakly-supervised medical image segmentation to generate pseudo segmentation masks with clearer boundaries and more accurate shapes by integrating two cause-effect chains including category-causality chain to alleviate the problem of ambiguous boundary and anatomy-causality chain to issue the co-occurrence challenge.
| Reference | 2D/3D | Modality | Dataset | Strategy to bridge the supervision gap |
|---|---|---|---|---|
| Zhang et al. [54] | 3D | MRI | ACDC [5], MSCMRseg [55, 56] | Maximize scribbles using mixed augmentation and random occlusion. |
| Luo et al. [57] | 3D | MRI | ACDC [5] | Randomly mixing outputs of dual-branch network to generate pseudo labels. |
| Asad et al. [58] | 3D | CT | UESTC-COVID-19 [59] | Extract patches exclusively with scribbles for online likelihood learning. |
| Yang et al. [60] | 3D | MRI | CHAOS [34], ACDC [5], LVSC [61] | Leverage a Siamese architecture with consistency training and entropy regularization. |
| Zhang et al. [62] | 3D | MRI | ACDC [5], MSCMRseg [55, 56] | Distinguish unlabeled pixels by maximizing the marginal probability. |
| Li et al. [63] | 3D | MRI | ACDC [5], MSCMRseg [55, 56], NCI-ISBI [33] | Use gated conditional random field loss for emphasizing boundaries between regions. |
III-B Medical Image Segmentation with Bounding Boxes
Bounding-box annotations can provide valuable information to guide models to identify and localize regions of interest, so using bounding-box annotations for segmentation is a popular topic in the field of medical imaging. Compared with image-level supervision, bounding box-level supervision serves is a more powerful form of annotation to narrow down the search space and enclose the segmented region within a rectangle, which offers a more comprehensive and informative representation of the target region’s location and extent and enables more accurate and robust segmentation.
To utilize weak annotations, pseudo masks are generated based on the annotated bounding boxes as priors. After that, a segmentation model is trained based on the pseudo masks. The primary difficulty in weakly supervised medical image segmentation with bounding boxes lies in accurately distinguishing foreground objects from the background regions within the bounding box. Rajchl et al. [35] propose DeepCut to achieve weakly supervised medical image segmentation based on bounding box annotations. Specifically, the segmentation problem is formulated as an energy minimization problem over a densely connected conditional random field with an iterative optimization to obtain pixel-wise segmentation from MRI images. Wang et al. [37] introduce a method for weakly supervised image segmentation that leverages tight bounding box annotations and utilizes generalized multiple instance learning (MIL) and smooth maximum approximation to integrate the bounding box tightness prior into a deep neural network in an end-to-end manner. Redekop et al. [39] develop a new bounding box correction framework to improve the tightness of non-tight 3D bounding box annotations used as weak labels, showing that this correction significantly enhances the performance of weakly supervised segmentation approaches. Wang et al. [64] present a novel weakly supervised approach for accurate segmentation of the prostate and nearby organs in CT images by utilizing 3D bounding box annotations and incorporating a label denoising module into the iterative training scheme to gradually refine the segmentation results. Afshari et al. [65] propose to utilize bounding boxes around tumor lesions for automatic tumor delineation in clinical PET scans by introducing a novel loss function that combines supervised and unsupervised components. Liu et al. [66] propose a weakly supervised learning method to utilize bounding box annotations by generating pseudo masks and incorporating data pre-processing techniques for segmenting liver, spleen, and kidney from CT images. Zhang et al. [67] introduce a weakly-supervised teacher-student network (WSTS) that utilizes additional box-level-labeled data to accurately segment liver tumors in non-enhanced images. Tang et al. [41] present a novel weakly-supervised universal lesion segmentation method based on HRNet [68] with a regional level set (RLS) loss for optimizing lesion boundary delineation and scale attention mechanisms to extract high-resolution deep image features crucial for accurate lesion segmentation. The RLS loss enables reliable and effective optimization in a weakly-supervised fashion, improving segmentation near lesion boundaries.
| Reference | 2D/3D | Modality | Dataset | Strategy to bridge the supervision gap |
|---|---|---|---|---|
| Kexrvadec et al. [69] | 3D | MRI | LV [5], VB [5], Prostate segmentation [70] | Enforce inequality constraints with differentiable penalties. |
| Zhai et al. [71] | 2D 3D | MRI | VS [72], BraTS2019 [73] | Contextual regularization using conditional random field and variance minimization and cross knowledge distillation. |
| Buhmann et al. [74] | 3D | Microscope | CREMI 1110 | Compare predicted synaptic connections with real synaptic point annotations. |
| Breznik et al. [75] | 2D | MRI | ACDC [5], POEM [76] | Calculate boundary loss using distance maps derived from point annotations. |
| Tian et al. [77] | 2D | Microscope | MoNuSeg[78], TNBC [79] | Integrate sparse supervised learning from points to regions and impose contour-sensitive constraints. |
1. https://cremi.org/
III-C Medical Image Segmentation with Scribble Annotations
Scribble is a powerful form of sparse annotation that can provide shape and size information for objects with complex shapes, as shown in Fig. 3. Scribble annotations are created by marking certain regions of an image with scribbles, which serve as guidance for the segmentation algorithm to accurately classify and delineate different objects or regions within the image. By requiring minimal input from users, scribble-based segmentation significantly reduces the manual effort required for detailed image labeling while still producing high-quality results. Consequently, this approach has practical applications in various fields such as medical imaging, scene understanding, object recognition, and video analysis. The strength of scribble segmentation lies in its ability to combine human intuition with computational power, resulting in more precise and efficient image segmentation.
Moreover, weakly supervised medical image segmentation based on scribble typically can involve leveraging the provided scribbles to guide the segmentation process. One common approach is to incorporate the scribble information into a loss function that encourages the segmentation model to produce predictions that align with the indicated rough outlines or hints. This can be achieved by leveraging the scribble annotations to set constraints for boundary areas in the loss function and guide models to calculate the unknown class directly. Asad et al. [58] present an efficient convolutional neural network (CNN) approach and use weighted cross-entropy loss to address the class imbalance that may result from user interactions. Zhang et al. [62] propose ShapePU based on the Positive-Unlabeled (PU) learning framework and global consistency regularization, leveraging unlabeled pixels via PU learning and exploiting shape knowledge. Can et al. [80] explore training strategies for learning the parameters of a pixel-wise segmentation network solely from scribble annotations. Dorent et al. [81] point out that scribbles on the target domain are used to perform domain adaptation with a new formulation of domain adaptation based on structured learning and co-segmentation. Wang et al. [82] enhance the framework by integrating CNNs into a pipeline for bounding box and scribble-based segmentation and adjusting parameters to make a CNN model adaptable to a specific test image.
Another approach is expanding scribble into the complete annotation, which utilizes foreground masking techniques to indicate or employs a background mask to indicate, which areas belong to the foregrounds. Lin et al. [83] show a weakly-supervised method for semantic segmentation based on scribbles, optimizing a graphical model for propagating information from scribbles. Xu et al. [84] reveal a Progressive Segmentation Inference (PSI) framework to tackle scribble-supervised semantic segmentation, which encapsulates two crucial cues, contextual pattern propagation, and semantic label diffusion. Luo et al. [57] utilize a dual-branch network comprising a single encoder and two marginally distinct decoders for image segmentation and blend the predictions from both decoders to create pseudo labels for auxiliary supervision. Zhang et al. [54] propose a novel weakly supervised segmentation framework, CycleMix that combines mix augmentation and cycle consistency. It adopts the mixup strategy with a specially designed random occlusion to perform increments and decrements of scribbles. Zhang et al. [85] exhibit to utilize the sole scribble annotations, investigating the principle of ”good scribble annotations”, which leads to efficient scribble forms via supervision maximization and randomness simulation, introducing regularization terms to encode the spatial relationship and shape prior, integrating the efficient scribble supervision with the prior into a unified framework. Li et al. [63] introduce a novel framework for scribble-supervised medical image segmentation that leverages vision and class embeddings via the multimodal information enhancement mechanism, utilizing both CNN features and transformer features uniformly to achieve enhanced visual feature extraction. Yang et al. [60] present the non-interactive method named PacingPseudo by comparing two weight-sharing networks, designing entropy regularization, distorted augmentations, and a new memory bank mechanism that provides an extra source of ensemble features to complement scarce labeled pixels. Zhuang et al. [86] give a training method only scribble guidance in the difficult areas. It uses a small set of fully annotated data to train the segmentation network and generates pseudo labels. Human supervisors draw scribbles in the areas of incorrect pseudo labels, and the scribbles are converted into pseudo label maps using a probability-modulated geodesic transform. Lee et al. [87] introduce Scribble2Label only hands-less scribble annotations and combines pseudo-labeling and label filtering to generate reliable labels from weak supervision.
III-D Medical Image Segmentation with Point Annotations
Point annotations are created by marking specific points of interest or landmarks on an image. These points are represented by coordinates indicating their positions within the image, as shown in Fig. 3. This annotation technique is commonly used for tasks like object detection, key point localization, facial landmark detection, and pose estimation. Point annotations can provide precise information about the locations of important features or objects, enabling algorithms to make accurate predictions and perform detailed analysis. When dense pixel-wise annotations are lack, weakly supervised segmentation with point annotation can be an effective method that uses auxiliary information or heuristic methods to estimate object or feature locations for annotation. Common methods include region-level annotation, weak label annotation, and auxiliary information annotation.
The region-level annotation work mainly involves drawing bounding boxes and assigning labels. Zhai et al. [88] propose a two-stage weakly supervised learning framework called PA-Seg for annotating segmentation in 3D medical images. The annotator only needs to provide seven points: one inside the target object as a foreground seed and six outside the target as background seeds. Breznik et al. [75] investigate the combination of intensity-based distance maps with boundary loss for point-supervised semantic segmentation, where the boundary loss penalizes false positives farther away from the object more strongly. The weak label annotation method uses fuzzy or partially labeled data for annotation. Yoo et al. [89] introduce PseudoedgeNet, a weakly supervised nuclei segmentation method that only requires point annotations for training. Gao et al. [90] manifest a framework that employs Minimal Point-Based annotation to accurately detect cancerous regions. The annotator only needs to mark a few cancerous and non-cancerous points in each whole-slide image (WSI). Buhmann et al. [74] propose a 3D U-Net architecture to identify pairs of voxels that are previous and postsynaptic to each other, formulating the problem of synaptic partner identification and allowing to directly learn from synaptic point annotations. Due to annotation uncertainty, results may introduce some noise or error. Therefore, caution is advised when using weakly supervised point annotation for training and analysis, considering the accuracy and reliability of the results.
In addition, several methods have been proposed as cascaded frameworks or coarse-to-fine frameworks to address the problem of limited or sparse annotations. Qu et al. [91] propose a weakly supervised nuclei segmentation framework that utilizes partial points annotation to train a detection model in the first stage and a segmentation model in the second stage, achieving competitive performance with significantly less annotation effort compared to fully supervised methods. Kexrvadec et al. [69] propose a weakly supervised segmentation framework that uses inequality constraints with a differentiable term to enforce constraints directly in the loss function, which has the potential to bridge the gap between weakly and fully supervised learning. Zhai et al. [71] indicate a method to annotate a segmentation target with only seven points, using geodesic distance transform in the first stage and leveraging model predictions as pseudo labels in the second stage. Tian et al. [77] express a coarse-to-fine weakly-supervised framework that employs self-stimulated learning through a self-supervision strategy using clustering for binary classification. Roth et al. [92, 93] propose to speed up medical image annotation by using minimal user interaction in the form of extreme point clicks to train a segmentation model, which is refined through multiple rounds of training and custom-designed loss and attention mechanism.
| Reference | 2D/3D | Modality | Dataset | Strategy to bridge the supervision gap |
|---|---|---|---|---|
| Zhang et al. [94] | 3D | CT | LiTS [95], KiTS [6], MSD [40] | Use a single network with a dynamic segmentation head to train from partially labeled datasets. |
| Dmitriev et al. [96] | 3D | CT | Sliver07 [97], NIH Pancreas [98] | Incorporate conditional information to implicitly share all parameters among target classes. |
| Zhou et al. [99] | 3D | CT | BTCV [100] | Incorporate anatomical priors to approximate empirical organ size distributions. |

III-E Medical Image Segmentation with Partially-Supervised Datasets
The workflow for medical image segmentation with partially-supervised datasets involves several steps. Firstly, the medical images and their corresponding annotations are collected and preprocessed. Then, the dataset is split into a fully-supervised subset and a partially-supervised subset. Next, a fully-supervised segmentation model is trained using the fully-supervised subset. Using this model, pseudo labels are generated for the partially-supervised subset. Subsequently, a segmentation model is trained on the combined fully-supervised and pseudo-labeled partially-supervised subsets, employing weakly-supervised or semi-supervised methods. To further enhance segmentation accuracy, the model is fine-tuned using the fully-supervised subset. Finally, the model is evaluated on a separate test set to validate its performance. This workflow enables accurate medical image segmentation even with limited labeled data by leveraging both fully and partially supervised learning techniques.
Zhang et al. [94] propose a dynamic on demand-network (DoDNet) which learns to segment multiple organs and tumors on partially labeled datasets. DoDNet consists of a shared encoder-decoder architecture, a task encoding module, a controller for dynamic filter generation, and a single but dynamic segmentation head. The task’s information from current segmentation tasks is encoded as a task-aware prior, guiding the model on the expected goals and outcomes of the task. Dmitriev et al. [96] present a unified highly efficient framework for robust simultaneous learning of multi-class segmentation by combining single-class datasets and utilizing a novel way of conditioning a convolutional network for segmentation. Zhou et al. [99] propose Prior-aware Neural Network (PaNN) to address the background ambiguity in partially-labeled datasets, which incorporates anatomical priors on abdominal organ sizes and guides the training process with domain-specific knowledge. Fidon et al. [101] advocate the first axiomatic definition of label-set loss functions which are the loss functions that can handle partially segmented images, proving that there is one and only one method to convert a classical loss function for fully segmented images into a proper label-set loss function. Huang et al. [102] design a novel general-purpose semi-supervised, multiple-task model—namely, self-supervised, semi-supervised, multitask learning (S4MTL) in medical imaging, segmentation, and diagnostic classification.
III-F Medical Image Segmentation with Mixed Supervision
Mix supervised learning in weakly supervised medical image segmentation addresses the problem of limited or incomplete annotations by combining different types of supervision signals. It leverages both weak annotations, such as scribbles or points, and strong annotations, such as fully segmented images, to improve the accuracy and robustness of the segmentation model. By combining these different sources of supervision, mixed supervised learning allows the model to learn from both coarse-level guidance and fine-level details, resulting in more accurate and detailed segmentations. This approach helps mitigate the challenges associated with weakly supervised segmentation and enhances the overall performance of the segmentation model in medical imaging tasks. This strategy is particularly important in cases where high-quality labeled data is scarce or expensive to obtain because it allows models to learn from weaker forms of supervision and still achieve high accuracy. Overall, mixed supervision has significant implications for the advancement of machine learning and artificial intelligence by enabling more efficient and effective learning from diverse data sources.
Mix supervised learning in weakly supervised medical image segmentation involves collecting a dataset with both weakly annotated images and strongly annotated images, designing a neural network architecture capable of handling both types of supervision signals, and training the network using a combination of weak and strong annotations. The loss function typically includes both weak and strong supervision terms, encouraging the network to learn from coarse-level guidance and fine-level details. By leveraging multiple sources of supervision, mix supervised learning improves the accuracy and robustness of medical image segmentation models, enabling the network to produce more precise and reliable segmentations. To address the challenge of limited annotated datasets for deep neural networks (DNNs) in medical image segmentation, Liu et al. [103] proposes a dual-branch architecture with a novel formulation incorporating Shannon entropy loss and Kullback-Leibler divergence to leverage mixed supervision, achieving substantial performance improvements compared to other strategies and recent semi-supervised approaches.
In order to address the high demand for accurate annotations in pixel-wise segmentation tasks, Rei et al. [104] present a semi-weakly supervised segmentation algorithm that utilizes deep supervision and a student-teacher model, achieving significant reduction in the requirement for expensive labels and narrowing the performance gap to fully supervised approaches using different types of annotations. Utilizing deep supervision and a student-teacher model allows for easy integration of different supervision signals. By carefully integrating deep supervision in lower layers and employing multi-label deep supervision, the authors achieve significant improvements in segmentation performance. Through a novel training regime that effectively uses various levels of annotation. Sun et al. [105] develop a semi-supervised learning framework based on a teacher-student fashion, propose a hierarchical organ-to-lesion (O2L) attention module in a teacher segmentor to produce pseudo-labels and train a student segmentor with combinations of manual- labeled and pseudo-labeled annotations.
By leveraging both weak and strong annotations and designing an appropriate loss function, mixed supervised learning enables the network to learn from different levels of supervision and improve the accuracy and robustness of medical image segmentation models. Shah et al. [106] propose a new FCN named MS-Net to reduce supervision cost by coupling strong supervision with weak supervision through low-cost input in the form of bounding boxes and landmarks. Shen et al. [107] give a mixed-supervision guided method and a residual-aided classification U-Net model (ResCU-Net) for joint segmentation and benign-malignant classification, which coupling the strong supervision in the form of segmentation mask and weak supervision in the form of benign-malignant label. Dolz et al. [108] indicate a dual-branch architecture. The upper branch teacher receives strong annotations but the bottom branch student is driven by limited supervision and guided by the upper branch, encouraging confident student predictions at the bottom branch and transferring the knowledge from the predictions generated by the strongly supervised branch to the less-supervised branch. Upadhyay et al. [109] show a novel generative adversarial gan network that can leverage training data at multiple levels of quality to improve performance while limiting the costs of data acquisition. Bhalgat et al. [110] present a budget-based cost-minimization framework in a mixed-supervision setting via dense segmentation, bounding boxes, and landmarks. The linear programming combines uncertainty with a similarity-based ranking strategy to select annotated examples.
IV Revisiting Weak Annotations in the Era of Foundation Models
The emergence of large-scale foundation models [111, 112] has revolutionized artificial intelligence (AI) and sparked a new era, primarily due to their remarkable zero-shot and few-shot generalization abilities across a wide range of downstream tasks [113]. Foundation models not only hold significant potential in natural language processing but have also broadened their application scope into computer vision with the emergence of vision-based foundation models [114, 115].
One pioneer example of vision foundation models [116] is the Segment Anything Model (SAM) [117], which has gained significant attention due to its impressive performance on a variety of semantic segmentation tasks [118, 119]. The design of SAM focuses on versatility, operational efficiency, and the inherent ability to handle ambiguity, representing significant progress in this field. It integrates an image encoder, a prompt encoder, and a mask decoder to generate masks for all objects within an image, demonstrating unprecedented zero-shot generalization for unseen objects or tasks. SAM’s versatility is evident in its ability to handle various types of prompts [116] such as points, bounding boxes, and free-form text, making it adaptable for various segmentation tasks. This adaptability is achieved through a combination of positional encodings for points and bounding boxes [120] and the complex integration of different embeddings for each type of cue, thereby enhancing its ability to effectively interpret various segmentation tasks. Significantly, the architecture of SAM allows it to produce multiple masks for a single prompt, effectively handling ambiguous cases [117]. This finding points to a significant aspect of vision foundation models: the utilization of weak annotations as prompts, which is fundamentally driven by the need for models to efficiently handle ambiguity and generalize from limited information. By effectively processing these simple prompts, SAM can adapt to various tasks and scenarios with increased efficiency, showing its versatility in diverse applications. This strategy represents a significant shift towards more adaptable and resource-efficient AI models.
In the domain of medical image segmentation with weak annotations, the application of SAM signifies a pivotal shift towards more efficient and scalable methodologies [121]. This shift is evidenced by a collection of studies that aim to optimize the annotation process and enhance the performance of segmentation models, leveraging minimal yet effective inputs like bounding boxes and point prompts. The evolution of these methods showcases a concerted effort to address the inherent challenges of medical image segmentation. Zhang et al. [122] indicate that using SAM with bounding box annotations to train segmentation models has high cost-effectiveness. Compared to models trained with pixel-wise annotations, this method shows competitive segmentation performance without the high demand for fully supervised annotations. This approach can provide a scalable and effective approach for medical image segmentation, potentially revolutionizing the annotation process and model training in this field. Building on this foundation, Deng et al. [123] further evaluate SAM’s performance in a more focused context, specifically in segmenting tumors, non-tumor tissues, and cell nuclei from whole slide imaging (WSI) utilizing both point annotations (both single and multiple) and bounding box annotations for annotation. They identify SAM’s remarkable performance in segmenting large connected objects but also uncover limitations in dense instance segmentation, pointing to the need for model fine-tuning and improved prompt selection.
To adapt to the nuanced demands of specific segmentation tasks and the challenge of effectively utilizing weak annotations, several studies have introduced tailored modifications to SAM, indicating its versatility and robustness in addressing these specialized needs. Feng et al. [124] employ image synthesis from a few exemplars to augment the dataset, and Low-Rank Adaptation (LoRA) [125] for fine-tuning SAM. This method demonstrates promising results in segmenting medical images with weak annotations, particularly in tasks like brain tumor segmentation and multi-organ CT segmentation, without specifying the exact type of weak annotations used for initial exemplar generation. In addition, Cui et al. [126] propose a pipeline that utilizes SAM, called all-in-SAM, for cell nucleus segmentation from bounding boxes to pixel-level labels, significantly exceeding the results obtained by supervised learning methods. This indicates that SAM has a strong ability to handle weak annotations of bounding boxes, and can clearly detect the nuclear boundaries within the focus area. Similarly, Wang et al. [127] introduce , a framework for medical image annotation using SAM, which consists of and . Among them, not only significantly improves the accuracy of SAM for medical image segmentation with only about five input points, but also combines points and bounding boxes to simulate real-world annotation scenes, demonstrating the adaptability of SAM.
Since SAM was originally designed for 2D natural images, it faces limitations in effectively extracting 3D spatial information from volumetric medical data [128]. To address these limitations of SAM, Wu et al. [129] propose a novel framework called Medical SAM Adapter (Med-SA) to enhance SAM’s ability to use weak annotations for high-quality medical image segmentation. Med-SA significantly expands SAM’s applicability in various medical imaging modes by introducing Space-Depth Transpose (SD-Trans) and Hyper-Prompting Adapter (HyP-Adpt). SD-Trans applies 2D SAM to 3D medical images, while HyP-Adpt allows prompt conditional model adaptation, enabling the model to effectively utilize click prompts and bounding boxes to generate accurate segmentation masks. Moreover, Gong et al. [130] also develop a novel method, 3DSAM-adapter, to extend the application of SAM from 2D to 3D images in medical data. This method adapts to 3D medical data by modifying the image encoder, prompt encoder, and mask decoder, effectively utilizing spatial context to improve segmentation performance. This adaptive improvement demonstrates superiority on four common tumor segmentation datasets using single-point annotations per volume, highlighting the potential and efficiency of using vision foundation models to handle weak annotations in the field of medical image segmentation.
SAM modes have been further improved by incorporating auxiliary modules or other software to capture additional contextual information. Liu et al. [131] describe the Segment Any Medical Model, an extension of SAM integrated into 3D Slicer [132] for medical image segmentation with weak annotations. This method involves the use of prompt points as annotations, where users interactively place points on the image slices to indicate regions of interest or to exclude certain areas. These prompt points are then used by the model to generate segmentation masks. Similarly, Lei et al. [133] develop MedLSAM, a model for automatic medical image segmentation that integrates a few-shot localization model [134] with the segmentation prowess of SAM for medical image segmentation. The localization model identifies anatomical regions across CT scans using self-supervision, reducing reliance on extensive manual annotations. It generates bounding boxes for image slices, which SAM utilizes for precise segmentation. This approach leverages weak annotations, such as minimal bounding boxes, simplifying segmentation tasks and enhancing the efficiency of processing limited labeled medical imagery. Zhang et al. [135] further adopt prompt augmentation of generated bounding box prompts for uncertainty estimation and rectification to enhance the reliability of segmentation results. Shahabany et al. [136] present AutoSAM, an improved SAM model specifically designed for medical image segmentation tasks. AutoSAM incorporates an auxiliary prompt encoder to directly use input images as prompts, rather than relying on manual weak annotations that SAM originally needed. This method enables AutoSAM to automatically adapt to the characteristics of medical images, improve segmentation performance, and reduce dependence on complex manual annotations. AutoSAM expands the application scope of SAM, making it more suitable for medical image segmentation tasks while maintaining the characteristics of weakly supervised learning. Chen et al. [137] explore the further applicability of SAM within the realm of weakly supervised segmentation by integrating text inputs and zero-shot learning settings. In the text input scenario, SAM employs class labels as text prompts, in combination with Grounded DINO [138] to generate bounding boxes, which are then used to produce segmentation masks. In the zero-shot learning scenario, the Recognize Anything Model (RAM) [139] is leveraged to identify class labels in the absence of explicit annotations, followed by a similar process of generating bounding boxes and masks. This method promotes medical image segmentation by explaining different prompts and demonstrating the potential of basic models in effectively processing weakly annotated data.
In addition to SAM and its variants, several researches also focus on other visual foundation models to expand the scope of medical image segmentation. Li et al. [140] propose ProMISe, a 3D medical image segmentation model using only a single point prompt. ProMISe utilizes point annotation to enable the model to focus on specific regions, improving accuracy and robustness in complex medical image segmentation tasks. Du et al. [141] introduce SegVol, a universal and interactive volumetric medical image segmentation foundation model. SegVol employs semantic and spatial prompts, including image-level annotations, bounding boxes, and point annotations, for precise segmentation through weak annotations. This model combines pseudo labels generated from unlabeled data with weakly supervised learning from a small amount of labeled datasets. Numerous experiments have demonstrated that SegVol outperforms state-of-the-art models on multiple segmentation benchmarks, particularly emphasizing its effectiveness in lesion segmentation on challenging datasets.
Collectively, these studies underscore the paradigm shift towards leveraging weak annotations and pioneering adaptations of vision foundation models for medical image segmentation. They highlight the continuous innovation in this field, enhancing the efficiency and applicability of segmentation models. This body of work paves the way for more scalable, annotation-effective solutions in medical imaging, promising significant advancements in the accuracy and efficiency of medical diagnoses and treatments.
V Discussion and Conclusion
The rapid advancements in the field of medical image segmentation, particularly with the advent of deep learning techniques, have significantly improved the accuracy and efficiency of automatically delineating anatomical structures or lesions from medical images. To ease the heavy reliance on pixel-wise annotations, which are labor-intensive and require expert knowledge, weakly supervised learning and the emergence of foundation models have opened new avenues for training deep models with limited or imperfect annotations, thus addressing the challenges associated with manual annotation. The weak supervision strategies have demonstrated the potential to reduce the reliance on extensive manual annotations while still achieving competitive segmentation performance. These methods leverage different forms of weak labels to guide the learning process, from high-level image-level labels to coarse scribbles and point locations, enabling the models to learn from available, yet limited weak annotations.
By combining the analysis of the latest literature, the following challenges are worthy of further discussion.
1) Quality evaluation and control of weak annotations. Assessing the quality of weak annotations is a critical challenge in weakly-supervised medical image segmentation. The performance of the segmentation model is heavily dependent on the quality of the weak annotations, which lack the granularity and precision compared with pixel-wise annotations [85]. Moreover, weak annotations come in various forms, and the disparity in quality across these different types can significantly impact the model’s performance. Weak annotations may originate from multiple sources including different annotators, annotation tools, or guidelines, and may contain noise, such as incorrect labels or inaccurate bounding boxes. These ambiguities may burden the development and lead to indecisiveness in the model during segmentation.
To address this challenge, there are several possible solutions. Firstly, the integration of various types of weak annotations, or combining weakly supervised learning with semi-supervised learning can enhance the contextual information for the model. Besides, it is important to develop specific metrics to quantify annotation quality, such as assessing consistency among different annotations or calculating overlap with a known ”gold standard” [142]. Furthermore, incorporating probabilistic models and graph learning techniques can be beneficial, as they can account for uncertainties in annotations and better capture the complex relationships within medical images.
2) Integrating domain knowledge. For medical image segmentation tasks, some organs have intricate structures that are difficult to segment accurately without pixel-wise supervision. For example, intra-kidney variability requires precise delineation that weakly-supervised methods may struggle to achieve due to the lack of fine-grained annotations [143]. Integrating domain knowledge can be a possible solution to overcome data scarcity and enhance the generalization capabilities and interpretability of segmentation model. For image data, one way is to explore prior information like position constraints and anatomical priors.
3) Utilizing existing datasets. Utilizing existing external datasets can help improve model generalization ability, which may come from different medical laboratories or related diseases [144]. These data may contain diverse information and enable the model to learn more knowledge specific to the medical field. The recent introduction of vision foundation models like the Segment Anything Model [26] has also shown the ability empowered by large-scale pre-training from natural images, with application to medical image segmentation tasks with weak annotations [145]. These models have also expanded the scope of medical image segmentation by incorporating weak annotations more effectively.
In conclusion, the survey presented in the document highlights the significant progress in medical image segmentation with weak annotations. The shift from traditional models to foundation models marks a paradigm change towards more flexible and scalable approaches that can generalize from limited information. The weakly supervised learning techniques have proven to be effective in reducing the annotation burden while maintaining high segmentation accuracy, which is crucial for clinical applications where expert time is valuable and resources are limited. The future of medical image segmentation lies in the continued development and refinement of these weakly supervised methods, as well as the integration of foundation models that can leverage weak annotations for improved performance. With the ongoing advancements in artificial general intelligence, it is expected that these models will become increasingly adept at handling ambiguous and limited data, further revolutionizing the field of medical imaging and contributing to more accurate diagnoses and effective treatments. As the research community continues to explore the capabilities of foundation models and weakly supervised learning, it is also crucial to focus on the practical applications and the potential impact on healthcare. The ultimate goal is to develop models that can seamlessly integrate into clinical workflows, providing radiologists and medical professionals with powerful tools to enhance the quality and speed of medical diagnostics. The path forward involves collaborative efforts between researchers, clinicians, and AI developers to ensure that these technologies are not only technologically advanced but also practically relevant and beneficial to the medical community.
References
- [1] G. Litjens, T. Kooi, B. E. Bejnordi, A. A. A. Setio, F. Ciompi, M. Ghafoorian, J. A. Van Der Laak, B. Van Ginneken, and C. I. Sánchez, “A survey on deep learning in medical image analysis,” Medical image analysis, vol. 42, pp. 60–88, 2017.
- [2] C. J. Lynch and C. Liston, “New machine-learning technologies for computer-aided diagnosis,” Nature Medicine, vol. 24, pp. 1304–1305, 2018.
- [3] J. Ma, Y. Zhang, S. Gu, C. Zhu, C. Ge, Y. Zhang, X. An, C. Wang, Q. Wang, X. Liu, S. Cao, Q. Zhang, S. Liu, Y. Wang, Y. Li, J. He, and X. Yang, “Abdomenct-1k: Is abdominal organ segmentation a solved problem?” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 44, no. 10, pp. 6695–6714, 2022.
- [4] I. Qureshi, J. Yan, Q. Abbas, K. Shaheed, A. B. Riaz, A. Wahid, M. W. J. Khan, and P. Szczuko, “Medical image segmentation using deep semantic-based methods: A review of techniques, applications and emerging trends,” Information Fusion, vol. 90, pp. 316–352, 2023.
- [5] O. Bernard, A. Lalande, C. Zotti, F. Cervenansky, X. Yang, P.-A. Heng, I. Cetin, K. Lekadir, O. Camara, M. A. G. Ballester et al., “Deep learning techniques for automatic mri cardiac multi-structures segmentation and diagnosis: is the problem solved?” IEEE transactions on medical imaging, vol. 37, no. 11, pp. 2514–2525, 2018.
- [6] N. Heller, F. Isensee, K. H. Maier-Hein, X. Hou, C. Xie, F. Li, Y. Nan, G. Mu, Z. Lin, M. Han et al., “The state of the art in kidney and kidney tumor segmentation in contrast-enhanced ct imaging: Results of the kits19 challenge,” Medical image analysis, vol. 67, p. 101821, 2021.
- [7] A. Lalande, Z. Chen, T. Pommier, T. Decourselle, A. Qayyum, M. Salomon, D. Ginhac, Y. Skandarani, A. Boucher, K. Brahim et al., “Deep learning methods for automatic evaluation of delayed enhancement-mri. the results of the emidec challenge,” Medical Image Analysis, vol. 79, p. 102428, 2022.
- [8] Ö. Çiçek, A. Abdulkadir, S. S. Lienkamp, T. Brox, and O. Ronneberger, “3d u-net: learning dense volumetric segmentation from sparse annotation,” in International conference on medical image computing and computer-assisted intervention. Springer, 2016, pp. 424–432.
- [9] O. Ronneberger, P. Fischer, and T. Brox, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, pp. 234–241.
- [10] X. Li, H. Chen, X. Qi, Q. Dou, C.-W. Fu, and P.-A. Heng, “H-denseunet: hybrid densely connected unet for liver and tumor segmentation from ct volumes,” IEEE transactions on medical imaging, vol. 37, no. 12, pp. 2663–2674, 2018.
- [11] Y. Zhang, L. Yuan, Y. Wang, and J. Zhang, “Sau-net: efficient 3d spine mri segmentation using inter-slice attention,” in Medical Imaging With Deep Learning. PMLR, 2020, pp. 903–913.
- [12] Z. Zhou, M. M. R. Siddiquee, N. Tajbakhsh, and J. Liang, “Unet++: Redesigning skip connections to exploit multiscale features in image segmentation,” IEEE transactions on medical imaging, vol. 39, no. 6, pp. 1856–1867, 2019.
- [13] J. Yang, P. Qiu, Y. Zhang, D. S. Marcus, and A. Sotiras, “D-net: Dynamic large kernel with dynamic feature fusion for volumetric medical image segmentation,” arXiv preprint arXiv:2403.10674, 2024.
- [14] F. Isensee, P. F. Jaeger, S. A. Kohl, J. Petersen, and K. H. Maier-Hein, “nnu-net: a self-configuring method for deep learning-based biomedical image segmentation,” Nature Methods, vol. 18, no. 2, pp. 203–211, 2021.
- [15] A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai, T. Unterthiner, M. Dehghani, M. Minderer, G. Heigold, S. Gelly et al., “An image is worth 16x16 words: Transformers for image recognition at scale,” in International Conference on Learning Representations, 2020.
- [16] J. Chen, Y. Lu, Q. Yu, X. Luo, E. Adeli, Y. Wang, L. Lu, A. L. Yuille, and Y. Zhou, “Transunet: Transformers make strong encoders for medical image segmentation,” arXiv preprint arXiv:2102.04306, 2021.
- [17] Y. Xie, J. Zhang, C. Shen, and Y. Xia, “Cotr: Efficiently bridging cnn and transformer for 3d medical image segmentation,” in International conference on medical image computing and computer-assisted intervention. Springer, 2021, pp. 171–180.
- [18] H.-Y. Zhou, J. Guo, Y. Zhang, X. Han, L. Yu, L. Wang, and Y. Yu, “nnformer: Volumetric medical image segmentation via a 3d transformer,” IEEE Transactions on Image Processing, 2023.
- [19] C. Jin, Z. Guo, Y. Lin, L. Luo, and H. Chen, “Label-efficient deep learning in medical image analysis: Challenges and future directions,” arXiv preprint arXiv:2303.12484, 2023.
- [20] V. M. Campello, P. Gkontra, C. Izquierdo, C. Martin-Isla, A. Sojoudi, P. M. Full, K. Maier-Hein, Y. Zhang, Z. He, J. Ma et al., “Multi-centre, multi-vendor and multi-disease cardiac segmentation: the m&ms challenge,” IEEE Transactions on Medical Imaging, vol. 40, no. 12, pp. 3543–3554, 2021.
- [21] Y. Zhang, Q. Liao, L. Ding, and J. Zhang, “Bridging 2d and 3d segmentation networks for computation-efficient volumetric medical image segmentation: An empirical study of 2.5 d solutions,” Computerized Medical Imaging and Graphics, p. 102088, 2022.
- [22] R. Jiao, Y. Zhang, L. Ding, B. Xue, J. Zhang, R. Cai, and C. Jin, “Learning with limited annotations: a survey on deep semi-supervised learning for medical image segmentation,” Computers in Biology and Medicine, p. 107840, 2023.
- [23] X. Luo, G. Wang, W. Liao, J. Chen, T. Song, Y. Chen, S. Zhang, D. N. Metaxas, and S. Zhang, “Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency,” Medical Image Analysis, vol. 80, p. 102517, 2022.
- [24] Y. Zhang, R. Jiao, Q. Liao, D. Li, and J. Zhang, “Uncertainty-guided mutual consistency learning for semi-supervised medical image segmentation,” Artificial Intelligence in Medicine, vol. 138, p. 102476, 2023.
- [25] Y. Shi, Y. Zhang, and S. Wang, “Competitive ensembling teacher-student framework for semi-supervised left atrium mri segmentation,” in 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). IEEE, 2023, pp. 2231–2234.
- [26] A. Kirillov, E. Mintun, N. Ravi, H. Mao, C. Rolland, L. Gustafson, T. Xiao, S. Whitehead, A. C. Berg, W.-Y. Lo et al., “Segment anything,” arXiv preprint arXiv:2304.02643, 2023.
- [27] Z. Jia, X. Huang, I. Eric, C. Chang, and Y. Xu, “Constrained deep weak supervision for histopathology image segmentation,” IEEE transactions on medical imaging, vol. 36, no. 11, pp. 2376–2388, 2017.
- [28] Y. Xu, J.-Y. Zhu, I. Eric, C. Chang, M. Lai, and Z. Tu, “Weakly supervised histopathology cancer image segmentation and classification,” Medical image analysis, vol. 18, no. 3, pp. 591–604, 2014.
- [29] X. Feng, J. Yang, A. F. Laine, and E. D. Angelini, “Discriminative localization in cnns for weakly-supervised segmentation of pulmonary nodules,” in Medical Image Computing and Computer Assisted Intervention- MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, September 11-13, 2017, Proceedings, Part III 20. Springer, 2017, pp. 568–576.
- [30] S. G. Armato III, G. McLennan, L. Bidaut, M. F. McNitt-Gray, C. R. Meyer, A. P. Reeves, B. Zhao, D. R. Aberle, C. I. Henschke, E. A. Hoffman et al., “The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans,” Medical physics, vol. 38, no. 2, pp. 915–931, 2011.
- [31] Z. Chen, Z. Tian, J. Zhu, C. Li, and S. Du, “C-cam: Causal cam for weakly supervised semantic segmentation on medical image,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2022, pp. 11 676–11 685.
- [32] G. Litjens, R. Toth, W. Van De Ven, C. Hoeks, S. Kerkstra, B. Van Ginneken, G. Vincent, G. Guillard, N. Birbeck, J. Zhang et al., “Evaluation of prostate segmentation algorithms for mri: the promise12 challenge,” Medical image analysis, vol. 18, no. 2, pp. 359–373, 2014.
- [33] K. Clark, B. Vendt, K. Smith, J. Freymann, J. Kirby, P. Koppel, S. Moore, S. Phillips, D. Maffitt, M. Pringle et al., “The cancer imaging archive (tcia): maintaining and operating a public information repository,” Journal of digital imaging, vol. 26, pp. 1045–1057, 2013.
- [34] A. E. Kavur, N. S. Gezer, M. Barış, S. Aslan, P.-H. Conze, V. Groza, D. D. Pham, S. Chatterjee, P. Ernst, S. Özkan et al., “Chaos challenge-combined (ct-mr) healthy abdominal organ segmentation,” Medical Image Analysis, vol. 69, p. 101950, 2021.
- [35] M. Rajchl, M. C. Lee, O. Oktay, K. Kamnitsas, J. Passerat-Palmbach, W. Bai, M. Damodaram, M. A. Rutherford, J. V. Hajnal, B. Kainz et al., “Deepcut: Object segmentation from bounding box annotations using convolutional neural networks,” IEEE transactions on medical imaging, vol. 36, no. 2, pp. 674–683, 2016.
- [36] M. S. Damodaram, L. Story, E. Eixarch, P. Patkee, A. Patel, S. Kumar, and M. Rutherford, “Foetal volumetry using magnetic resonance imaging in intrauterine growth restriction,” Early human development, vol. 88, pp. S35–S40, 2012.
- [37] J. Wang and B. Xia, “Bounding box tightness prior for weakly supervised image segmentation,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part II. Springer, 2021, pp. 526–536.
- [38] S.-L. Liew, J. M. Anglin, N. W. Banks, M. Sondag, K. L. Ito, H. Kim, J. Chan, J. Ito, C. Jung, N. Khoshab et al., “A large, open source dataset of stroke anatomical brain images and manual lesion segmentations,” Scientific data, vol. 5, no. 1, pp. 1–11, 2018.
- [39] E. Redekop and A. Chernyavskiy, “Medical image segmentation with imperfect 3d bounding boxes,” in Deep Generative Models, and Data Augmentation, Labelling, and Imperfections: First Workshop, DGM4MICCAI 2021, and First Workshop, DALI 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, October 1, 2021, Proceedings 1. Springer, 2021, pp. 193–200.
- [40] M. Antonelli, A. Reinke, S. Bakas, K. Farahani, A. Kopp-Schneider, B. A. Landman, G. Litjens, B. Menze, O. Ronneberger, R. M. Summers et al., “The medical segmentation decathlon,” Nature Communications, vol. 13, no. 1, pp. 1–13, 2022.
- [41] Y. Tang, J. Cai, K. Yan, L. Huang, G. Xie, J. Xiao, J. Lu, G. Lin, and L. Lu, “Weakly-supervised universal lesion segmentation with regional level set loss,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part II 24. Springer, 2021, pp. 515–525.
- [42] K. Yan, X. Wang, L. Lu, and R. M. Summers, “Deeplesion: automated mining of large-scale lesion annotations and universal lesion detection with deep learning,” Journal of medical imaging, vol. 5, no. 3, pp. 036 501–036 501, 2018.
- [43] B. Zhou, A. Khosla, À. Lapedriza, A. Oliva, and A. Torralba, “Learning deep features for discriminative localization,” 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2921–2929, 2015.
- [44] R. R. Selvaraju, A. Das, R. Vedantam, M. Cogswell, D. Parikh, and D. Batra, “Grad-cam: Visual explanations from deep networks via gradient-based localization,” International Journal of Computer Vision, vol. 128, pp. 336–359, 2016.
- [45] A. Chattopadhyay, A. Sarkar, P. Howlader, and V. N. Balasubramanian, “Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks,” 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), pp. 839–847, 2017.
- [46] H. Wang, Z. Wang, M. Du, F. Yang, Z. Zhang, S. Ding, P. P. Mardziel, and X. Hu, “Score-cam: Score-weighted visual explanations for convolutional neural networks,” 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), pp. 111–119, 2019.
- [47] M. Früh, M. Fischer, A. Schilling, S. Gatidis, and T. Hepp, “Weakly supervised segmentation of tumor lesions in pet-ct hybrid imaging,” Journal of Medical Imaging, vol. 8, no. 5, pp. 054 003–054 003, 2021.
- [48] Y. Li, Y. Liu, L. Huang, Z. Wang, and J. Luo, “Deep weakly-supervised breast tumor segmentation in ultrasound images with explicit anatomical constraints,” Medical image analysis, vol. 76, p. 102315, 2022.
- [49] M. Izadyyazdanabadi, E. Belykh, C. Cavallo, X. Zhao, S. Gandhi, L. B. Moreira, J. Eschbacher, P. Nakaji, M. C. Preul, and Y. Yang, “Weakly-supervised learning-based feature localization for confocal laser endomicroscopy glioma images,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part II 11. Springer, 2018, pp. 300–308.
- [50] S. Hwang and H.-E. Kim, “Self-transfer learning for weakly supervised lesion localization,” in Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, October 17-21, 2016, Proceedings, Part II 19. Springer, 2016, pp. 239–246.
- [51] J. J. Yoo, K. Namdar, and F. Khalvati, “Superpixel generation and clustering for weakly supervised brain tumor segmentation in mr images,” arXiv preprint arXiv:2209.09930, 2022.
- [52] F. Dubost, H. Adams, P. Yilmaz, G. Bortsova, G. van Tulder, M. A. Ikram, W. Niessen, M. W. Vernooij, and M. de Bruijne, “Weakly supervised object detection with 2d and 3d regression neural networks,” Medical Image Analysis, vol. 65, p. 101767, 2020.
- [53] V. N. Dang, F. Galati, R. Cortese, G. Di Giacomo, V. Marconetto, P. Mathur, K. Lekadir, M. Lorenzi, F. Prados, and M. A. Zuluaga, “Vessel-captcha: an efficient learning framework for vessel annotation and segmentation,” Medical Image Analysis, vol. 75, p. 102263, 2022.
- [54] K. Zhang and X. Zhuang, “Cyclemix: A holistic strategy for medical image segmentation from scribble supervision,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2022, pp. 11 656–11 665.
- [55] X. Zhuang, “Multivariate mixture model for cardiac segmentation from multi-sequence mri,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2016, pp. 581–588.
- [56] ——, “Multivariate mixture model for myocardial segmentation combining multi-source images,” IEEE transactions on pattern analysis and machine intelligence, vol. 41, no. 12, pp. 2933–2946, 2018.
- [57] X. Luo, M. Hu, W. Liao, S. Zhai, T. Song, G. Wang, and S. Zhang, “Scribble-supervised medical image segmentation via dual-branch network and dynamically mixed pseudo labels supervision,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2022: 25th International Conference, Singapore, September 18–22, 2022, Proceedings, Part I. Springer, 2022, pp. 528–538.
- [58] M. Asad, L. Fidon, and T. Vercauteren, “Econet: Efficient convolutional online likelihood network for scribble-based interactive segmentation,” in International Conference on Medical Imaging with Deep Learning. PMLR, 2022, pp. 35–47.
- [59] G. Wang, X. Liu, C. Li, Z. Xu, J. Ruan, H. Zhu, T. Meng, K. Li, N. Huang, and S. Zhang, “A noise-robust framework for automatic segmentation of covid-19 pneumonia lesions from ct images,” IEEE Transactions on Medical Imaging, vol. 39, no. 8, pp. 2653–2663, 2020.
- [60] Z. Yang, D. Lin, D. Ni, and Y. Wang, “Non-iterative scribble-supervised learning with pacing pseudo-masks for medical image segmentation,” Expert Systems with Applications, vol. 238, p. 122024, 2024.
- [61] A. Suinesiaputra, B. R. Cowan, A. O. Al-Agamy, M. A. Elattar, N. Ayache, A. S. Fahmy, A. M. Khalifa, P. Medrano-Gracia, M.-P. Jolly, A. H. Kadish et al., “A collaborative resource to build consensus for automated left ventricular segmentation of cardiac mr images,” Medical image analysis, vol. 18, no. 1, pp. 50–62, 2014.
- [62] K. Zhang and X. Zhuang, “Shapepu: A new pu learning framework regularized by global consistency for scribble supervised cardiac segmentation,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2022: 25th International Conference, Singapore, September 18–22, 2022, Proceedings, Part VIII. Springer, 2022, pp. 162–172.
- [63] Z. Li, Y. Zheng, X. Luo, D. Shan, and Q. Hong, “Scribblevc: Scribble-supervised medical image segmentation with vision-class embedding,” in Proceedings of the 31st ACM International Conference on Multimedia, 2023, pp. 3384–3393.
- [64] S. Wang, Q. Wang, Y. Shao, L. Qu, C. Lian, J. Lian, and D. Shen, “Iterative label denoising network: Segmenting male pelvic organs in ct from 3d bounding box annotations,” IEEE Transactions on Biomedical Engineering, vol. 67, no. 10, pp. 2710–2720, 2020.
- [65] S. Afshari, A. BenTaieb, Z. Mirikharaji, and G. Hamarneh, “Weakly supervised fully convolutional network for pet lesion segmentation,” in Medical imaging 2019: image Processing, vol. 10949. SPIE, 2019, pp. 394–400.
- [66] Y. Liu, Q. Hui, Z. Peng, S. Gong, and D. Kong, “Automatic ct segmentation from bounding box annotations using convolutional neural networks,” arXiv preprint arXiv:2105.14314, 2021.
- [67] D. Zhang, B. Chen, J. Chong, and S. Li, “Weakly-supervised teacher-student network for liver tumor segmentation from non-enhanced images,” Medical Image Analysis, vol. 70, p. 102005, 2021.
- [68] J. Wang, K. Sun, T. Cheng, B. Jiang, C. Deng, Y. Zhao, D. Liu, Y. Mu, M. Tan, X. Wang, W. Liu, and B. Xiao, “Deep high-resolution representation learning for visual recognition,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 43, pp. 3349–3364, 2019.
- [69] H. Kervadec, J. Dolz, M. Tang, E. Granger, Y. Boykov, and I. B. Ayed, “Constrained‐cnn losses for weakly supervised segmentation,” Medical Image Analysis, vol. 54, p. 88–99, 2018.
- [70] C. Chu, D. L. Belavỳ, G. Armbrecht, M. Bansmann, D. Felsenberg, and G. Zheng, “Fully automatic localization and segmentation of 3d vertebral bodies from ct/mr images via a learning-based method,” PloS one, vol. 10, no. 11, p. e0143327, 2015.
- [71] S. Zhai, G. Wang, X. Luo, Q. Yue, K. Li, and S. Zhang, “Pa-seg: Learning from point annotations for 3d medical image segmentation using contextual regularization and cross knowledge distillation,” IEEE Transactions on Medical Imaging, 2023.
- [72] J. Shapey, G. Wang, R. Dorent, A. Dimitriadis, W. Li, I. Paddick, N. Kitchen, S. Bisdas, S. R. Saeed, S. Ourselin et al., “An artificial intelligence framework for automatic segmentation and volumetry of vestibular schwannomas from contrast-enhanced t1-weighted and high-resolution t2-weighted mri,” Journal of neurosurgery, vol. 134, no. 1, pp. 171–179, 2019.
- [73] B. H. Menze, A. Jakab, S. Bauer, J. Kalpathy-Cramer, K. Farahani, J. Kirby, Y. Burren, N. Porz, J. Slotboom, R. Wiest et al., “The multimodal brain tumor image segmentation benchmark (brats),” IEEE transactions on medical imaging, vol. 34, no. 10, pp. 1993–2024, 2014.
- [74] J. Buhmann, R. Krause, R. C. Lentini, N. Eckstein, M. Cook, S. Turaga, and J. Funke, “Synaptic partner prediction from point annotations in insect brains,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part II 11. Springer, 2018, pp. 309–316.
- [75] E. Breznik, H. Kervadec, F. Malmberg, J. Kullberg, H. Ahlström, M. de Bruijne, and R. Strand, “Leveraging point annotations in segmentation learning with boundary loss,” arXiv preprint arXiv:2311.03537, 2023.
- [76] L. Lind, “Relationships between three different tests to evaluate endothelium-dependent vasodilation and cardiovascular risk in a middle-aged sample,” Journal of hypertension, vol. 31, no. 8, pp. 1570–1574, 2013.
- [77] K. Tian, J. Zhang, H. Shen, K. Yan, P. Dong, J. Yao, S. Che, P. Luo, and X. Han, “Weakly-supervised nucleus segmentation based on point annotations: A coarse-to-fine self-stimulated learning strategy,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part V 23. Springer, 2020, pp. 299–308.
- [78] N. Kumar, R. Verma, S. Sharma, S. Bhargava, A. Vahadane, and A. Sethi, “A dataset and a technique for generalized nuclear segmentation for computational pathology,” IEEE transactions on medical imaging, vol. 36, no. 7, pp. 1550–1560, 2017.
- [79] P. Naylor, M. Laé, F. Reyal, and T. Walter, “Segmentation of nuclei in histopathology images by deep regression of the distance map,” IEEE transactions on medical imaging, vol. 38, no. 2, pp. 448–459, 2018.
- [80] Y. B. Can, K. Chaitanya, B. Mustafa, L. M. Koch, E. Konukoglu, and C. F. Baumgartner, “Learning to segment medical images with scribble-supervision alone,” in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4. Springer, 2018, pp. 236–244.
- [81] R. Dorent, S. Joutard, J. Shapey, S. Bisdas, N. Kitchen, R. Bradford, S. Saeed, M. Modat, S. Ourselin, and T. Vercauteren, “Scribble-based domain adaptation via co-segmentation,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part I 23. Springer, 2020, pp. 479–489.
- [82] G. Wang, W. Li, M. A. Zuluaga, R. Pratt, P. A. Patel, M. Aertsen, T. Doel, A. L. David, J. Deprest, S. Ourselin et al., “Interactive medical image segmentation using deep learning with image-specific fine tuning,” IEEE transactions on medical imaging, vol. 37, no. 7, pp. 1562–1573, 2018.
- [83] D. Lin, J. Dai, J. Jia, K. He, and J. Sun, “Scribblesup: Scribble-supervised convolutional networks for semantic segmentation,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 3159–3167.
- [84] J. Xu, C. Zhou, Z. Cui, C. Xu, Y. Huang, P. Shen, S. Li, and J. Yang, “Scribble-supervised semantic segmentation inference,” in Proceedings of the IEEE/CVF International Conference on Computer Vision, 2021, pp. 15 354–15 363.
- [85] K. Zhang and X. Zhuang, “Zscribbleseg: Zen and the art of scribble supervised medical image segmentation,” arXiv preprint arXiv:2301.04882, 2023.
- [86] M. Zhuang, Z. Chen, Y. Yang, L. Kettunen, and H. Wang, “Annotation-efficient training of medical image segmentation network based on scribble guidance in difficult areas,” International Journal of Computer Assisted Radiology and Surgery, pp. 1–10, 2023.
- [87] H. Lee and W.-K. Jeong, “Scribble2label: Scribble-supervised cell segmentation via self-generating pseudo-labels with consistency,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part I 23. Springer, 2020, pp. 14–23.
- [88] S. Zhai, G. Wang, X. Luo, Q. Yue, K. Li, and S. Zhang, “Pa-seg: Learning from point annotations for 3d medical image segmentation using contextual regularization and cross knowledge distillation,” IEEE transactions on medical imaging, 2022.
- [89] I. Yoo, D. Yoo, and K. Paeng, “Pseudoedgenet: Nuclei segmentation only with point annotations,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part I 22. Springer, 2019, pp. 731–739.
- [90] Z. Gao, P. Puttapirat, J. Shi, and C. Li, “Renal cell carcinoma detection and subtyping with minimal point-based annotation in whole-slide images,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part V 23. Springer, 2020, pp. 439–448.
- [91] H. Qu, P. Wu, Q. Huang, J. Yi, Z. Yan, K. Li, G. M. Riedlinger, S. De, S. Zhang, and D. N. Metaxas, “Weakly supervised deep nuclei segmentation using partial points annotation in histopathology images,” IEEE Transactions on Medical Imaging, vol. 39, pp. 3655–3666, 2020.
- [92] H. Roth, L. Zhang, D. Yang, F. Milletari, Z. Xu, X. Wang, and D. Xu, “Weakly supervised segmentation from extreme points,” in Large-Scale Annotation of Biomedical Data and Expert Label Synthesis and Hardware Aware Learning for Medical Imaging and Computer Assisted Intervention: International Workshops, LABELS 2019, HAL-MICCAI 2019, and CuRIOUS 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13 and 17, 2019, Proceedings 4. Springer, 2019, pp. 42–50.
- [93] H. R. Roth, D. Yang, Z. Xu, X. Wang, and D. Xu, “Going to extremes: weakly supervised medical image segmentation,” Machine Learning and Knowledge Extraction, vol. 3, no. 2, pp. 507–524, 2021.
- [94] J. Zhang, Y. Xie, Y. Xia, and C. Shen, “Dodnet: Learning to segment multi-organ and tumors from multiple partially labeled datasets,” in Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, 2021, pp. 1195–1204.
- [95] P. Bilic, P. Christ, H. B. Li, E. Vorontsov, A. Ben-Cohen, G. Kaissis, A. Szeskin, C. Jacobs, G. E. H. Mamani, G. Chartrand et al., “The liver tumor segmentation benchmark (lits),” Medical Image Analysis, vol. 84, p. 102680, 2023.
- [96] K. Dmitriev and A. E. Kaufman, “Learning multi-class segmentations from single-class datasets,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2019, pp. 9501–9511.
- [97] T. Heimann, B. Van Ginneken, M. A. Styner, Y. Arzhaeva, V. Aurich, C. Bauer, A. Beck, C. Becker, R. Beichel, G. Bekes et al., “Comparison and evaluation of methods for liver segmentation from ct datasets,” IEEE transactions on medical imaging, vol. 28, no. 8, pp. 1251–1265, 2009.
- [98] R. Holger and F. Amal, “Turkbey evrim, lu le, liu jiamin, and summers ronald. data from pancreas–ct,” Cancer Imaging Archive, 2016.
- [99] Y. Zhou, Z. Li, S. Bai, C. Wang, X. Chen, M. Han, E. Fishman, and A. L. Yuille, “Prior-aware neural network for partially-supervised multi-organ segmentation,” in Proceedings of the IEEE/CVF international conference on computer vision, 2019, pp. 10 672–10 681.
- [100] B. Landman, Z. Xu, J. Igelsias, M. Styner, T. Langerak, and A. Klein, “Miccai multi-atlas labeling beyond the cranial vault–workshop and challenge,” in Proc. MICCAI Multi-Atlas Labeling Beyond Cranial Vault—Workshop Challenge, vol. 5, 2015, p. 12.
- [101] L. Fidon, M. Aertsen, S. Shit, P. Demaerel, S. Ourselin, J. Deprest, and T. Vercauteren, “Partial supervision for the feta challenge 2021,” arXiv preprint arXiv:2111.02408, 2021.
- [102] C. Huang, H. Tang, W. Fan, Y. Xiao, D. Hao, Z. Qian, D. Terzopoulos et al., “Partly supervised multi-task learning,” in 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA). IEEE, 2020, pp. 769–774.
- [103] B. Liu, C. Desrosiers, I. B. Ayed, and J. Dolz, “Segmentation with mixed supervision: Confidence maximization helps knowledge distillation,” Medical Image Analysis, vol. 83, p. 102670, 2023.
- [104] S. Reiß, C. Seibold, A. Freytag, E. Rodner, and R. Stiefelhagen, “Every annotation counts: Multi-label deep supervision for medical image segmentation,” 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 9527–9537, 2021.
- [105] L. Sun, J. Wu, X. Ding, Y. Huang, G. Wang, and Y. Yu, “A teacher-student framework for semi-supervised medical image segmentation from mixed supervision,” arXiv preprint arXiv:2010.12219, 2020.
- [106] M. P. Shah, S. Merchant, and S. P. Awate, “Ms-net: mixed-supervision fully-convolutional networks for full-resolution segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2018, pp. 379–387.
- [107] T. Shen, C. Gou, J. Wang, and F.-Y. Wang, “Simultaneous segmentation and classification of mass region from mammograms using a mixed-supervision guided deep model,” IEEE Signal Processing Letters, vol. 27, pp. 196–200, 2019.
- [108] J. Dolz, C. Desrosiers, and I. B. Ayed, “Teach me to segment with mixed supervision: Confident students become masters,” in Information Processing in Medical Imaging: 27th International Conference, IPMI 2021, Virtual Event, June 28–June 30, 2021, Proceedings 27. Springer, 2021, pp. 517–529.
- [109] U. Upadhyay and S. P. Awate, “A mixed-supervision multilevel gan framework for image quality enhancement,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2019, pp. 556–564.
- [110] Y. Bhalgat, M. Shah, and S. Awate, “Annotation-cost minimization for medical image segmentation using suggestive mixed supervision fully convolutional networks,” arXiv preprint arXiv:1812.11302, 2018.
- [111] X. Wang, G. Chen, G. Qian, P. Gao, X.-Y. Wei, Y. Wang, Y. Tian, and W. Gao, “Large-scale multi-modal pre-trained models: A comprehensive survey,” Machine Intelligence Research, pp. 1–36, 2023.
- [112] P. P. Liang, A. Zadeh, and L.-P. Morency, “Foundations and recent trends in multimodal machine learning: Principles, challenges, and open questions,” arXiv preprint arXiv:2209.03430, 2022.
- [113] R. Bommasani, D. A. Hudson, E. Adeli, R. Altman, S. Arora, S. von Arx, M. S. Bernstein, J. Bohg, A. Bosselut, E. Brunskill et al., “On the opportunities and risks of foundation models,” arXiv preprint arXiv:2108.07258, 2021.
- [114] X. Wang, X. Zhang, Y. Cao, W. Wang, C. Shen, and T. Huang, “Seggpt: Segmenting everything in context,” arXiv preprint arXiv:2304.03284, 2023.
- [115] G. Wang, J. Wu, X. Luo, X. Liu, K. Li, and S. Zhang, “Mis-fm: 3d medical image segmentation using foundation models pretrained on a large-scale unannotated dataset,” arXiv preprint arXiv:2306.16925, 2023.
- [116] K.-H. Jung, “Uncover this tech term: foundation model,” Korean Journal of Radiology, vol. 24, no. 10, p. 1038, 2023.
- [117] A. Kirillov, E. Mintun, N. Ravi, H. Mao, C. Rolland, L. Gustafson, T. Xiao, S. Whitehead, A. C. Berg, W.-Y. Lo et al., “Segment anything,” arXiv preprint arXiv:2304.02643, 2023.
- [118] F. Li, H. Zhang, P. Sun, X. Zou, S. Liu, J. Yang, C. Li, L. Zhang, and J. Gao, “Semantic-sam: Segment and recognize anything at any granularity,” arXiv preprint arXiv:2307.04767, 2023.
- [119] J. Yang, M. Gao, Z. Li, S. Gao, F. Wang, and F. Zheng, “Track anything: Segment anything meets videos,” arXiv preprint arXiv:2304.11968, 2023.
- [120] M. Tancik, P. Srinivasan, B. Mildenhall, S. Fridovich-Keil, N. Raghavan, U. Singhal, R. Ramamoorthi, J. Barron, and R. Ng, “Fourier features let networks learn high frequency functions in low dimensional domains,” Advances in Neural Information Processing Systems, vol. 33, pp. 7537–7547, 2020.
- [121] Y. Zhang, Z. Shen, and R. Jiao, “Segment anything model for medical image segmentation: Current applications and future directions,” Computers in Biology and Medicine, 2024.
- [122] Y. Zhang, S. Zhao, H. Gu, and M. A. Mazurowski, “How to efficiently annotate images for best-performing deep learning based segmentation models: An empirical study with weak and noisy annotations and segment anything model,” 2023.
- [123] R. Deng, C. Cui, Q. Liu, T. Yao, L. W. Remedios, S. Bao, B. A. Landman, L. E. Wheless, L. A. Coburn, K. T. Wilson et al., “Segment anything model (sam) for digital pathology: Assess zero-shot segmentation on whole slide imaging,” arXiv preprint arXiv:2304.04155, 2023.
- [124] W. Feng, L. Zhu, and L. Yu, “Cheap lunch for medical image segmentation by fine-tuning sam on few exemplars,” arXiv preprint arXiv:2308.14133, 2023.
- [125] E. J. Hu, Y. Shen, P. Wallis, Z. Allen-Zhu, Y. Li, S. Wang, L. Wang, and W. Chen, “Lora: Low-rank adaptation of large language models,” arXiv preprint arXiv:2106.09685, 2021.
- [126] C. Cui, R. Deng, Q. Liu, T. Yao, S. Bao, L. W. Remedios, Y. Tang, and Y. Huo, “All-in-sam: from weak annotation to pixel-wise nuclei segmentation with prompt-based finetuning,” arXiv preprint arXiv:2307.00290, 2023.
- [127] C. Wang, D. Li, S. Wang, C. Zhang, Y. Wang, Y. Liu, and G. Yang, “Sam med: A medical image annotation framework based on large vision model,” arXiv preprint arXiv:2307.05617, vol. 3, 2023.
- [128] M. Awais, M. Naseer, S. Khan, R. M. Anwer, H. Cholakkal, M. Shah, M.-H. Yang, and F. S. Khan, “Foundational models defining a new era in vision: A survey and outlook,” arXiv preprint arXiv:2307.13721, 2023.
- [129] J. Wu, R. Fu, H. Fang, Y. Liu, Z. Wang, Y. Xu, Y. Jin, and T. Arbel, “Medical sam adapter: Adapting segment anything model for medical image segmentation,” arXiv preprint arXiv:2304.12620, 2023.
- [130] S. Gong, Y. Zhong, W. Ma, J. Li, Z. Wang, J. Zhang, P.-A. Heng, and Q. Dou, “3dsam-adapter: Holistic adaptation of sam from 2d to 3d for promptable medical image segmentation,” arXiv preprint arXiv:2306.13465, 2023.
- [131] Y. Liu, J. Zhang, Z. She, A. Kheradmand, and M. Armand, “Samm (segment any medical model): A 3d slicer integration to sam,” arXiv preprint arXiv:2304.05622, 2023.
- [132] A. Fedorov, R. Beichel, J. Kalpathy-Cramer, J. Finet, J.-C. Fillion-Robin, S. Pujol, C. Bauer, D. Jennings, F. Fennessy, M. Sonka et al., “3d slicer as an image computing platform for the quantitative imaging network,” Magnetic resonance imaging, vol. 30, no. 9, pp. 1323–1341, 2012.
- [133] W. Lei, X. Wei, X. Zhang, K. Li, and S. Zhang, “Medlsam: Localize and segment anything model for 3d medical images,” arXiv preprint arXiv:2306.14752, 2023.
- [134] W. Lei, W. Xu, R. Gu, H. Fu, S. Zhang, S. Zhang, and G. Wang, “Contrastive learning of relative position regression for one-shot object localization in 3d medical images,” in Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part II 24. Springer, 2021, pp. 155–165.
- [135] Y. Zhang, S. Hu, C. Jiang, Y. Cheng, and Y. Qi, “Segment anything model with uncertainty rectification for auto-prompting medical image segmentation,” arXiv preprint arXiv:2311.10529, 2023.
- [136] T. Shaharabany, A. Dahan, R. Giryes, and L. Wolf, “Autosam: Adapting sam to medical images by overloading the prompt encoder,” arXiv preprint arXiv:2306.06370, 2023.
- [137] Z. Chen and Q. Sun, “Weakly-supervised semantic segmentation with image-level labels: from traditional models to foundation models,” arXiv preprint arXiv:2310.13026, 2023.
- [138] S. Liu, Z. Zeng, T. Ren, F. Li, H. Zhang, J. Yang, C. Li, J. Yang, H. Su, J. Zhu et al., “Grounding dino: Marrying dino with grounded pre-training for open-set object detection,” arXiv preprint arXiv:2303.05499, 2023.
- [139] Y. Zhang, X. Huang, J. Ma, Z. Li, Z. Luo, Y. Xie, Y. Qin, T. Luo, Y. Li, S. Liu et al., “Recognize anything: A strong image tagging model,” arXiv preprint arXiv:2306.03514, 2023.
- [140] H. Li, H. Liu, D. Hu, J. Wang, and I. Oguz, “Promise: Prompt-driven 3d medical image segmentation using pretrained image foundation models,” arXiv preprint arXiv:2310.19721, 2023.
- [141] Y. Du, F. Bai, T. Huang, and B. Zhao, “Segvol: Universal and interactive volumetric medical image segmentation,” arXiv preprint arXiv:2311.13385, 2023.
- [142] D. Hartmann, V. Schmid, P. Meyer, I. Soto-Rey, D. Müller, and F. Kramer, “Mism: A medical image segmentation metric for evaluation of weak labeled data,” arXiv preprint arXiv:2210.13642, 2022.
- [143] H. Du, Q. Dong, Y. Xu, and J. Liao, “Weakly-supervised 3d medical image segmentation using geometric prior and contrastive similarity,” IEEE Transactions on Medical Imaging, 2023.
- [144] Y. Zhang, Q. Liao, L. Yuan, H. Zhu, J. Xing, and J. Zhang, “Exploiting shared knowledge from non-covid lesions for annotation-efficient covid-19 ct lung infection segmentation,” IEEE Journal of Biomedical and Health Informatics, vol. 25, no. 11, pp. 4152–4162, 2021.
- [145] Y. Zhao, T. Zhou, Y. Gu, Y. Zhou, Y. Zhang, Y. Wu, and H. Fu, “Segment anything model-guided collaborative learning network for scribble-supervised polyp segmentation,” arXiv preprint arXiv:2312.00312, 2023.