Assessment of contextualised representations in detecting outcome phrases in clinical trials
Abstract
Background: Automating the recognition of outcomes reported in clinical trials using machine learning has a huge potential of speeding up access to evidence necessary in healthcare decision making. Prior research has however acknowledged inadequate training corpora as a challenge for the Outcome detection (OD) task. Additionally, several contextualised representations (embeddings) like BERT and ELMO have achieved unparalleled success in detecting various diseases, genes, proteins and chemicals, however, the same cannot be emphatically stated for outcomes, because these representation models have been relatively under-tested and studied for the OD task.
Methods: We introduce “EBM-COMET”, a dataset in which 300 Randomised Clinical Trial (RCT) PubMed abstracts are expertly annotated for clinical outcomes. Unlike prior related datasets that use arbitrary outcome classifications, we use labels from a taxonomy recently published to standardise outcome classifications. To extract outcomes, we fine-tune a variety of pre-trained contextualised representations, additionally, we use frozen contextualised and context-independent representations in our custom neural model augmented with clinically informed Part-Of-Speech embeddings and a cost-sensitive loss function. We adopt strict evaluation for the trained models by rewarding them for correctly identifying full outcome phrases rather than words within the entities i.e. given an outcome phrase “systolic blood pressure”, the models are rewarded a classification score only when they predict all 3 words in sequence, otherwise, they are not rewarded.
Results and Conclusion: We observe our best model (BioBERT) achieve 81.5% F1, 81.3% sensitivity and 98.0% specificity. We reach a consensus on which contextualised representations are best suited for detecting outcome phrases from clinical trial abstracts. Furthermore, our best model outperforms scores published on the original EBM-NLP dataset leader-board scores.
Keywords: Outcome detection, Outcome dataset, Contextualised representations, Transfer Learning, Full outcome phrase.
1 Introduction
There is growing recognition of the potential benefits of using readily available sources of clinical information to support clinical research [?]. Of particular importance is the identification of information about outcomes measured on patients, for example, blood pressure, fatigue, etc. The ability to automatically detect outcome phrases contained within clinical narrative text will serve to maximise the potential of such sources. For example, hospital or GP letters, or free text fields recorded within electronic health records, may contain valuable clinical information which is not readily accessible or analysable without manual or automated extraction of relevant outcome phrases. Similarly, automated identification of outcomes mentioned in trial registry entries or trial publications could help to facilitate systematic review processes by speeding up outcome data extraction. Furthermore, the benefits of automated outcome recognition will be increased further if it extends to categorisation of outcomes within a relevant classification system such as taxonomy proposed in [?]. The potential contribution of Natural Language Processing (NLP) to EBM [?] has been limited by the scarcity of publicly available annotated corpora [?] and the inconsistency in how outcomes are described in different trials [?; ?; ?]. Nonetheless, rapid advancement in NLP techniques has accelerated NLP-powered EBM research, enabling tasks such as detecting elements that collectively form the basis of clinical questions including Participants/population (P), Interventions (I), Comparators (C), and Outcomes (O) [?]. I and C are often collapsed into just I [?; ?; ?].
EBM-NLP corpus [?] is the only publicly available corpus that can support individual outcome phrase detection. However, this dataset used an arbitrary selection of outcome classifications despite being aligned to Medical Subject Headings (MESH)111https://www.nlm.nih.gov/mesh/. Moreover, it contains flawed outcome annotations [?] such as measurement tools and statistical metrics incorrectly annotated as outcomes and others which we mention in section 2.
In this work, we are motivated by the outcome taxonomy recently built and published to standardise outcome classifications [?]. We work closely with experts to annotate outcomes with classification drawn from this taxonomy.
Several variations of state-of-the-art (SOTA) CLMs that include BioBERT [?], SciBERT [?], ClinicalBERT [?] and others have recently emerged to aid clinical NLP tasks. Despite their outstanding performance in multiple clinical NLP tasks such as BNER [?; ?] and relation extraction [?], they have been underutilised for the outcome detection task, mainly because of inadequate corpora [?]. Given that, clinical trial abstracts (which report outcomes) are part of the medical text on which these CLMs are pre-trained, we leverage transfer learning (TL) and make full use of them to achieve individual outcome detection. The goal in the outcome detection task is to extract outcome phrases from clinical text. For example, in a sentence,“Among patients who received sorafenib, the most frequently reported adverse events, were grade 1 or 2 events of rash (73%), fatigue 67%, hypertension (55%) and diarrhea (51%)”, we extract all outcome phrases such as those underlined and in bold font. This enables those searching the literature including patients and policymakers to identify research that addresses the health outcomes of most importance to them [?]. Following previous studies that investigated which embeddings are best suited for clinical-NLP text classification tasks [?], we focus this work on probing for some consensus amongst various SOTA domain-specific CLM embeddings, determining which embeddings are best suited for outcome detection. A summary of our contributions includes,
-
1.
We introduce a novel outcome dataset, EBM-COMET, in which outcomes within randomised clinical trial (RCT) abstracts are expertly annotated with outcome classifications drawn from [?].
-
2.
We assess the performance of domain-specific (clinical) context-dependent representations in comparison to generic context-dependent and context-independent representations for the outcome detection task.
-
3.
We assess the quality in detecting full mention of outcome phrases in comparison to detection of individual words contained in outcome phrases. Ideally, given an outcome phrase “systolic blood pressure”, full outcome phrase evaluation strictly rewards models for correctly detecting all 3 words in that sequence (exact match), whereas word-level evaluation rewards models for correctly detecting any single word in phrase. The former is particularly informative for the biomedical domain audience [?].
-
4.
We compare the performance of the CLMs in our experimental setup to the current leader-board performance on extracting PIO elements from the original EBM-NLP dataset [?].
2 Related Work
2.1 Outcome detection
Outcome detection has previously been simultaneously achieved along with Participant and Intervention detection, where researchers aim to classify sentences (extracted from RCT abstracts) into one of P, I and O labels [?; ?; ?]. Despite being restrained by shortage of expertly labeled datasets, few attempts to create EBM-oriented datasets have been made. Bryon et al., [?] use distant supervision to annotate sentences within clinical trial articles with PICO elements. Dina et al., [?] use an experienced Nurse and a medical student to annotate outcomes by identifying and labelling sentences that best summarise the consequence of an intervention. Similarly, other attempts have precisely partitioned PubMed abstracts into sentences that they label one of P, I, and O [?; ?]. Given the sentence-level annotation adopted in these datasets, it becomes difficult to use them for tasks that require extraction of individual PICO elements [?; ?] such as outcome phrase detection. Nye et al., [?] recently released EBM-NLP corpus that they built using a mixture of crowd workers (non-experts) and expert workers (with the non-experts being exceedingly more) to annotate individual spans of P, I, O elements within clinical trial articles. This dataset has however been discovered with annotation flaws [?] and uses arbitrary outcome classification labels as discussed in section 2. Cognizant of the growing body of research to standardise classifications of outcomes, we are motivated to annotate a dataset with outcome types drawn from a standardised taxonomy.
2.2 Transfer Learning (TL)
TL is a machine learning (ML) approach that enables usage of a model to achieve a task that it was not initially built and trained for [?]. Usually, the assumption is that, train and test data for a specific task exists, however this is never the case, therefore, TL allows learning across different task domains i.e. the term pre-trained, implies a model was previously trained on a task different from the current target task. Context-dependent embeddings such as context2vec [?], ELMo [?] and BERT [?] have emerged and outperformed context-independent embeddings [?; ?] in various downstream NLP tasks.
Bert variants, SciBERT [?] and ClinicalBERT [?] yielded performance improvements in the BNER tasks on the BC5DR dataset [?; ?], text-classification tasks like Relation extraction on the ChemProt [?] and on PICO extraction. Despite being pre-trained on English biomedical text, BioBERT [?] outperformed generic BERT model ( pre-trained on Spanish biomedical text) in Pharma-CoNER, a multi-classification task for detecting mentions of chemical names and drugs from Spanish biomedical text [?]. Recently Qiao et al., [?] discovered that, in comparison to BioBERT, BioELMo (Biomedical ELMo) better clustered entities of the same type such as, an acronym having multiple meanings or a homonym. For example, unlike BioBERT, BioELMo clearly differentiated between ER referring to “Estrogen Receptor” and ER referring to “Emergency Room” in their work.
3 Materials and Methods
We design two setups in our assessment approach, where (1) we fine-tune pre-trained biomedical CLMs on the outcome datasets EBM-COMET (introduced in this paper) and EBM-NLP (a revised version of the original EBM-NLP [?]) and (2) we augment a neural model to train frozen biomedical embeddings. The aim is to compare the evaluation performance of fine-tuned, frozen biomedical CLM embeddings, generic CLM embeddings and traditional context-independent embeddings such as word2Vec [?] in the outcome detection task defined below.
Outcome Detection Problem (ODP) Task:
Given a sentence s of n words, within an RCT abstract, outcome detection aims to extract an outcome phrase within , where . In order to extract outcome phrases such as , we label each word using the “BIO” tagging scheme [?] where “B” denotes the first word of the outcome phrase, “I” denotes inside the outcome phrase and “O” denotes all non-outcome phrase words.
3.1 Data
EBM-COMET
EBM-COMET is prepared to facilitate outcome detection in EBM. Our annotation scheme adopts a widely acknowledged definition of outcome which is “a measurement or an observation used to capture and assess the effect of treatment such as assessment of side effects (risk) or effectiveness (benefits)” [?]. Previous EBM dataset construction efforts have lacked a standard classification system to accurately inform their annotation process and instead opted for arbitrary labels such as those terms aligned to MeSH [?]. We however leverage an outcome taxonomy recently developed to standardise outcome reporting in electronic databases [?]. The taxonomy authors iteratively reviewed how core outcome sets (COS) studies within the Core Outcome Measures in Effectiveness Trials (COMET) database categorised their outcomes. This review culminated into a taxonomy of 38 outcome domains hierarchically classified into 5 outcome types/core areas.
Data collection
Using the Entrez API [?], we automatically fetch 300 abstracts from open access PubMed. Our search criteria only retrieve articles of type “Randomised controlled Trial”. We relied on two domain-experts to review these abstracts and eliminate those reporting outcomes in animals (or non-humans). Each eliminated abstract was replaced by another reporting human outcomes from PubMed.
Annotation
The two experts we work with have sufficient experience in reviewing human health outcomes in clinical trials. Some of their work pertaining to outcomes in clinical trials includes [?; ?; ?; ?]. These experts jointly annotate granular outcomes within the gathered abstracts resulting into EBM-COMET using guidelines below. We are aware of annotation tools such as BRAT [?], however because of the nature of the annotations i.e. some with contiguous outcome spans, the experts prefer to directly annotate them in Microsoft text documents.
Annotation guidelines
The annotators are tasked to identify and verify outcome spans and then assign each an outcome domain referenced from the taxonomy partially presented in table 1 and full presented in Appendix C. The annotators are instructed to assign each span all relevant outcome domains.
Annotation heuristics
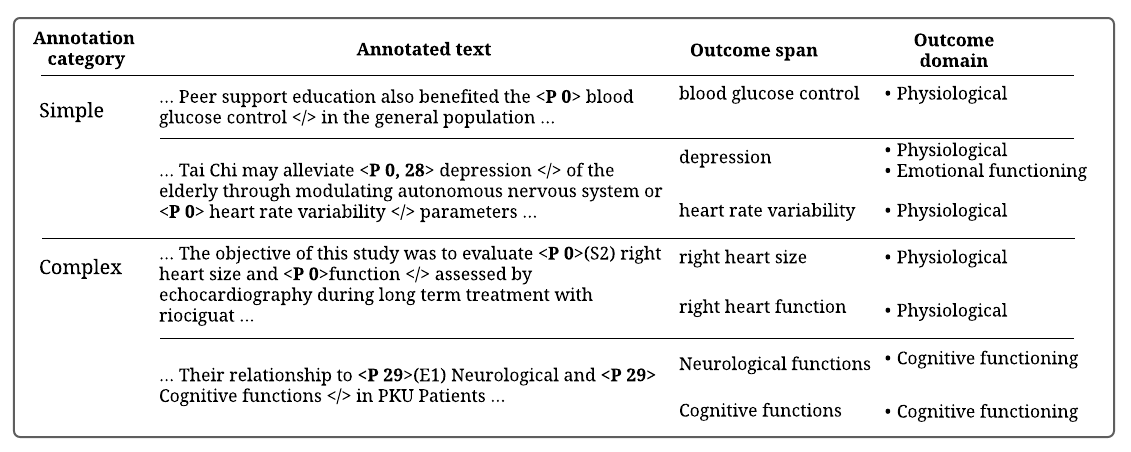
For annotation purposes, we firstly assign a unique symbol to each outcome domain (domain symbol column in table 1). The annotators are then instructed to use these symbols to label the outcome spans they identify. Annotation using these symbols rather than the long domain names is less tedious. Furthermore, we instruct annotators to use xml tags to demarcate the spans, such that an identified span is enclosed within an opening tag with the assigned domain symbol and a closing tag. We refer to easily identifiable outcome spans as simple annotations, and the more difficult ones requiring more demarcation indicators as complex annotations. Figure 1 shows examples of the annotations described below,
-
1.
Simple annotations
-
(a)
P XX…/: Indicates an outcome belongs to domain XX (where XX can be located in the taxonomy 1).
-
(b)
P XX, YY…/: Indicates an outcome belongs to both domain XX and YY.
-
(a)
-
2.
Complex annotations
Some spans are contiguous in such a way that, they share a word or words with other spans. For example, two outcomes can easily be annotated as a single outcome because they are conjoined by a dependency word or punctuation such as “and”, “or” and commas. We are however fully aware, that this contiguity previously resulted in multiple outcomes annotated as a single outcome in previous datasets [?]. Therefore, annotators are asked to distinctively annotate them as below,-
(a)
Contiguous spans sharing bordering term/s appearing at the start of an outcome span should be annotated as follows,
P XX(S#)…P XX…/: which indicates that, two outcomes are belonging to domain XX that share # of words at the start of the annotated outcome span. -
(b)
Contiguous spans sharing bordering term/s appearing at the end of an outcome span, should be annotated as follows,
P XX(E#)…P XX…/: The opposite of the notation above indicating that, two outcomes are belonging to domain XX that share # of words at the end of the annotated outcome span.
-
(a)
Annotation consistency and quality
In the last phase of the annotation process, the annotations are extracted into a structured format (excel sheet) for the annotators to review them, make necessary alterations based on their expertise judgement as well as handle minor errors (such as wrong opening or closing braces) that result from the manual annotation processes. We do not report inter-annotator agreement because the two annotators did not conduct the process independently, but rather jointly. Having previously worked together on similar annotation tasks, they hardly disagreed but whenever either was uncertain or disagreed, they discussed between themselves and concluded.
The word, outcome phrase distribution and other statistics of the EBM-COMET are summarised in table 4 with the experimental dataset statistics.
| Core area | Outcome domain |
|
||
|---|---|---|---|---|
|
|
P 0 | ||
| Death | Mortality/survival | P 1 | ||
| Life Impact | Physical functioning | P 25 | ||
| Social functioning | P 26 | |||
| Role functioning | P 27 | |||
|
P 28 | |||
| Cognitive functioning | P 29 | |||
| Global quality of life | P 30 | |||
| Perceived health status | P 31 | |||
| Delivery of care | P 32 | |||
| Personal circumstances | P 33 | |||
| Resource use | Economic | P 34 | ||
| Hospital | P 35 | |||
|
P 36 | |||
| Societal/carer burden | P 37 | |||
|
|
P 38 |
EBM-NLP
This dataset is a revision of the original hierarchical label’s version of EBM-NLP dataset [?]. In the hierarchical labels version, the annotated outcome spans were assigned specific labels that include Physical, Pain, Mental, Mortality and Adverse effects. Abaho et al., [?] built EBM-NLP using a semi-automatic approach that involved POS-tagging and rule-based chunking to correct flaws discovered (by domain-experts) in EBM-NLP. In the evaluation of this revision, classification of outcomes resulted in a significant increase in the F1-score (for all labels) from what it was when using the original EBM-NLP. Some of the major flaws they corrected include,
-
•
Statistical metrics and measurement tools annotated as part of clinical outcomes e.g. “mean arterial blood-pressure” instead of “arterial blood-pressure”, “Quality of life Questionnaire” instead of “Quality of life”, “Work-related stress scores” instead of “Work-related stress”.
-
•
Multiple outcomes annotated as a single outcome e.g. “cardiovascular events-(myocardial infarction, stroke and cardiovascular death)” instead of “myocardial infarction”, “stroke”, and “cardiovascular death”.
-
•
Inaccurate outcome type annotations e.g. “Nausea and Vomiting” labeled as a Mortality outcome instead of a Physical outcome.
-
•
Combining annotations in non-human studies with those in human-studies particularly studies reporting outcomes in treating beef cattle.
3.2 Biomedical Contextual Language Models
We leverage the datasets to investigate the ODP task performance of 6 different biomedical CLMs (table 2) derived from 3 main architectures. 1) BERT [?], a CLM built by learning deep bidirectional representations of input words by jointly incorporating left and right context in all its layers. It works by masking a portion of the input words and thereby predicting missing words in each sentence. BERT encodes a word by incorporating information about words around it within a given input sentence using a self-attention mechanism [?] 2) ELMo [?] is a CLM that learns deep bidirectional representations of input words by jointly maximizing the probability of forward and backward directions in a sentence, and 3) FLAIR [?], a character-level bidirectional LM which learns representations of each character by incorporating character information around it within a sequence of words.
| Model |
|
Pre-trained on | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Bert | BioBERT [?] |
|
|||||||
| SciBERT [?] |
|
||||||||
| ClinicalBERT [?] |
|
||||||||
|
|
||||||||
| ELMo | BioELMo [?] |
|
|||||||
| FLAIR | BioFLAIR [?] | 1.8m PubMed abstracts. |

We begin by further training the pre-trained CLMs in table 2 in a fine-tuning approach [?], where the CLMs learn to (1) encode each word into a hidden state and (2) predict the correct label given . Similar to Sun et al.. [?], we introduce a non-linear softmax layer to predict a label for each corresponding to word , as shown in figure 2, where , {BERT-variants, BioELMo, BioFLAIR} . (see Appendix A.1 (Fine-tuning) for more details).
3.3 ODP-tagger
We build ODP-tagger to not only assess context-independent (W2V) representations, but also assess the performance of frozen context-dependent representations for the ODP task. Demonstrated by the dotted line from Fine-tuning to input tokens in figure 2, is a feature extraction [?] approach, where the tagger’s embedding layer takes as input, a sequence of tokens (sentence) and a sequence of POS terms corresponding to the tokens. We add a POS feature for each token to enrich the model in a manner similar to how prior neural classifiers are enhanced with character and n-gram features [?]. Each word/token is therefore represented by concatenating either a pre-trained CLM or a W2V embedding and a randomly initialised embedding for the corresponding POS term . The token embeddings are then encoded to obtain hidden-states for each sequence position,
| (1) |
where and , denote Word and POS matrices, each containing d-dimensional embeddings for words and corresponding POS terms. and are the word and POS embeddings representing the word and its POS term, ; implies a concatenation operation and then is a linear activation function that generates hidden states for the input words. We then use a condition random field (CRF) layer for classification given the hidden state . A CRF is an undirected graphical model which defines a conditional probability distribution over possible labels [?].
All the models are each trained to maximize the probability of the labels given each word .
| (2) |
The training loss objective.
| (3) |
where is a scaling factor that empirically sets each labels weights to be inversely proportional to the square root of the label frequency i.e. and is the number of training samples with ground-truth label . is the training set containing sentences, and .
| Fine-tuning | Feature extraction | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Model | EBM-NLP | EBM-COMET | Model | EBM-NLP | EBM-COMET | ||||
| W2V | _ | _ | ODP-tagger + W2V | 44.0 | 59.3 | ||||
| BERT | 51.8 | 75.5 | +BERT | 43.2 | 64.2 | ||||
| ELMO | 49.6 | 71.4 | +ELMO | 43.0 | 61.2 | ||||
| BioBERT | 53.1 | 81.5 | +BioBERT | 48.5 | 69.3 | ||||
| BioELMO | 52.0 | 75.0 | +BioELMO | 46.5 | 62.9 | ||||
| BioFLAIR | 51.4 | 76.7 | +BioFLAIR | 40.7 | 60.5 | ||||
| SciBERT | 52.8 | 77.6 | +SciBERT | 48.1 | 70.4 | ||||
| ClinicalBERT | 51.0 | 68.5 | +ClinicalBERT | 45.2 | 65.7 | ||||
| Bio+ClinicalBERT | 51.0 | 68.3 | +Bio+ClinicalBERT | 45.8 | 66.3 | ||||
|
51.0 | 70.0 |
|
46.1 | 68.4 | ||||
3.4 Training
All models are evaluated on the two datasets discussed in section 3.1. These datasets are each partitioned as follows, 75% for training (train), 15% for development (dev) and 10% for testing (test). We exploit the large size of EBM-NLP (as shown in table 4) and use its dev set to tune hyperparameters for the ODP-tagger and fine-tuned models (Parameter settings in Appendix B). Each model is trained on a train split of a particular dataset and evaluated on the corresponding test split culminating into results shown in table 3. We use a simple powerful NLP python framework called flair222https://github.com/flairNLP/flair to extract word embeddings from all the BERT and FLAIR variants, and AllenAI333https://github.com/allenai/bilm-tf for BioELMO. Dimensions of the extracted BioFLAIR and BioELMO embeddings are very large, i.e. 7672 and 3072 respectively, which would most likely overwhelm our memory and power-constrained devices during training. Therefore, we apply Principal component Analysis (PCA) dimensionality reduction technique to reduce their dimensions to half their original sizes while preserving semantic information [?]. Alongside these embeddings, we evaluate context-independent embeddings which we obtain by training word2vec (W2V) embedding algorithm [?] on 5.5B tokens of PubMed and PMC abstracts. Python and PyTorch [?] deep learning framework are used for implementation, which together with the datasets are made publicly available here https://github.com/MichealAbaho/ODP-tagger.
| EBM-COMET | EBM-NLP | ||||
| # of sentences | 5193 | 40092 | |||
|
3895 / 779 / 519 | 30069 / 6014 / 4009 | |||
| # of outcome labels | 5 | 6 | |||
|
1569 / 451 / 221 | 12481 / 4116 / 3257 | |||
|
20.6 / 21.5 / 21.2 | 25.5 / 26.4 / 25.6 | |||
|
0.69 / 0.78 / 0.71 | 0.44 / 0.38 / 0.45 |
3.5 Evaluation results
Results shown in table 3 firstly reveal the superiority of fine-tuning the CLMs in comparison to the ODP-tagger. The best performance across both set-ups is obtained when BioBERT is fine-tuned on the EBM-COMET dataset. However, we observe SciBERT outperform it in the ODP-tagger set-up on the EBM-COMET dataset. Secondly, we observe CLM embeddings produce stronger performances in comparison to context-independent (W2V) embeddings especially with the EBM-COMET dataset. BioFLAIR and ClinicalBERT were the least performing models. For BioFLAIR, we hypothesize that, (1) pre-training on a relatively smaller corpus, (2) it being of much less depth (1-layered BiLSTM) compared to multi-layered BERT and ELMo and (3) downsizing its embeddings using PCA dimensionality reduction are reasons that led to its low performance. For ClinicalBERT, we attributed its struggles to the nature of the corpora on which it is pre-trained. Unlike BioBERT, SciBERT and BioELMo which are pre-trained on PubMed text which is mostly clinical trial abstracts that more often report health outcomes, ClinicalBERT is pre-trained on clinical notes associated with patient hospital admissions [?]. An additional insight we drew was, performance on the EBM-NLP dataset is lower compared to that achieved on EBM-COMET. This was attributed to the annotation inconsistencies in the original EBM-NLP, some of which were resolved in [?]. Another aspect we closely observed was the runtime. Using a TITAN RTX 24GB GPU, the average runtime for the fine-tuning experiments on EBM-COMET and EBM-NLP respectively was 7 and 12 hrs. On the other-hand, feature extraction (ODP-tagger) experiments were much longer consuming 20 and 36 hours respectively on the same datasets. Overall, we recommend fine-tuning as a preferred approach for outcome detection, more saw using BioBERT and SciBERT as ideal embedding models.
| Method | Abstract sentence | Full outcome phrase | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||
|
Output |
|
- adverse events - rash | - fatigue - hypertension - diarrhea | |||||||||||
|
Output |
|
- fatigue - diarrhea | - hypertension | |||||||||||
|
|
- duration of operating procedure | |||||||||||||
|
Output |
|
|||||||||||||
|
Output |
|
|||||||||||||
|
|
|
|||||||||||||
|
Output |
|
- right heart size | ||||||||||||
|
Output |
|
|||||||||||||
3.6 Full outcome phrase detection
Motivated by the need to detect accurate fine-grained information in the medical domain [?], we examine the extent to which our models detect precise mentions of full outcome phrases. To achieve this, we investigate how well the best performing models (Fine-tuned+BioBERT+EBM-COMET and Fine-tuned+BioBERT+EBM-NLP from Table 3) can detect full mentions of outcome phrases or otherwise exact matches of outcome phrases in prediction results. We use a strict criteria to evaluate full mention of outcomes, where a classification error FN (False Negative) accounts for the number of full outcome phrases the model fails to detect, which includes partially correctly detected phrases i.e. some of their tokens were misclassified. In table 6, we observe the F1 of the best models drop from 53.1 to 52.4 for EBM-NLP and 81.5 to 69.6 for EBM-COMET. This implies that the model struggles to identify full outcome phrases, especially with the EBM-NLP dataset. Specificity on the other hand is very high for both datasets simply because it is calculated as a True Negative Rate (TNR), in which case True Negatives (non-outcomes) are certainly so many because they are precisely individual words and therefore are counted word by word as opposed to True positives (actual outcome phrases) that can consist of multiple words.
| P | R | S | F | |
|---|---|---|---|---|
| EBM-NLP | 53.7 | 51.2 | 99.2 | 52.4 |
| EBM-COMET | 60.8 | 81.3 | 98.0 | 69.6 |
We further investigate the errors from the best performing models BioBERT+EBM-COMET (Fine-tuned) and ODP-tagger+SciBERT+EBM-COMET. In table 5, we show examples of outputs of both models for the ODP task given an input sentence with known actual outcome phrases (underlined). Fine-tuned model correctly detects (blue-coded) all full outcome phrase in the first example sentence i.e. Precision (P), Recall/Sensitivity (R) are 100%, whereas tagger only detects 3/4 outcomes, hence P is 100%, R is 75%. Neither of the models correctly capture full mention of the outcome phrase in the second example, they incorrectly predict some words (red-coded) to not belong to the outcome phrase. While traditionally, results of fine-tuned model would be a P of 100% and R of 50% for correct prediction of 2/4 tokens, in our strict full name evaluation, P and R are 0%, because some tokens in the full outcome phrase are mis-classified in both models i.e. True positives = 0. Similarly, in the third example, fine-tuned model achieves P of 100% and R of 60% for correct prediction of 3/5 tokens in the traditional evaluation, whereas for the strict full name evaluation, R is 50% because only 1/2 full outcome phrases are detected. We attribute these errors to the length of some outcome phrases with some containing extremely common words such as prepositions (“of”). Additionally, we note that the contiguous outcome span annotations (containing several outcomes sharing terms e.g. “right heart size and function” in the third example) are rare.
3.7 Evaluation on the original EBM-NLP
We additionally fine-tune our best model for the task of detection of all PIO elements in the original EBM-NLP dataset. To be consistent with the original EBM-NLP paper, we consider the token-level detection of the PIO elements task in their work, comparing their evaluation results for hierarchical labels with those we obtain by fine-tuning our best model. Using their published training (4670) and test (190) sets of the starting spans, we see fine-tuned BioBERT model outperform the current leader board results 444https://ebm-nlp.herokuapp.com/ and the SOTA results published by Brockmeier et al [?] (table 7). We attribute this improvement to the fact, unlike the LSTM-CRF and Logreg models in previous SOTA scores, BioBERT’s has an internal capability to encode information using self-attention mechanisms to generate context-sensitive representations of words.
| P | I | O | |||
|---|---|---|---|---|---|
| Logreg | 45.0 | 25.0 | 38.0 | ||
| Lstm-crf | 40.0 | 50.0 | 48.0 | ||
| Brockmeier et.al [?] | 70.0 | 56.0 | 70.0 | ||
| Fine-tuned BioBERT | 71.6 | 69.0 | 73.1 | ||
|
61.6 | 64.0 | 53.1 |
3.8 Outcome phrase length
To further understand our results, we investigated how well the best models BioBERT+EBM-COMET (Fine-tuned) and ODP-tagger+SciBERT+EBM-COMET (Feature-extraction) detected outcome phrases of varying lengths. We calculate a prediction accuracy as number of correctly predicted outcome-phrases of length x/number of all outcome-phrases of length x, where x ranged from 1-10. As observed in figure 3, the fine-tuned model slightly outperforms the ODP-tagger especially for outcome phrases having 3-6 words (i.e. 3-6 entity span length). However, it is also clear that both models struggled to accurately detect outcome phrases containing 7 or more words.

4 Conclusion
In this work, we present EBM-COMET, a dataset of clinical trial abstracts with outcome annotations to facilitate EBM tasks. Experiments showed that CLMs perform much better on EBM-COMET than they do on EBM-NLP, indicating it is suited for ODP task especially because it is well aligned to standardised outcome classifications. Our assessment showed) fine-tuned models consistently outperform and converge faster than feature extraction, particularly pre-trained BioBERT and SciBERT embedding models. Additionally, we show the significance of accurate detection of full mention of granular outcome phrases which is beneficial for clinicians searching for this information.
References
- [1] Bartlett VL, Dhruva SS, Shah ND, Ryan P, Ross JS. Feasibility of Using Real-World Data to Replicate Clinical Trial Evidence [Journal Article]. JAMA network open. 2019;2(10):e1912869–e1912869.
- [2] Dodd S, Clarke M, Becker L, Mavergames C, Fish R, Williamson PR. A taxonomy has been developed for outcomes in medical research to help improve knowledge discovery. Journal of Clinical Epidemiology. 2018 4;96:84–92.
- [3] Sackett DL, Rosenberg WMC, Gray JAM, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn’t. BMJ. 1996;312(7023):71–72. Available from: https://www.bmj.com/content/312/7023/71.
- [4] Nye B, Li JJ, Patel R, Yang Y, Marshall IJ, Nenkova A, et al. A corpus with multi-level annotations of patients, interventions and outcomes to support language processing for medical literature. In: Proceedings of the conference. Association for Computational Linguistics. Meeting; 2018. p. 197.
- [5] Coiera E, Choong MK, Tsafnat G, Hibbert P, Runciman WB. Linking quality indicators to clinical trials: an automated approach [Journal Article]. International Journal for Quality in Health Care. 2017;29(4):571–578.
- [6] Demner-Fushman D, Lin J. Answering clinical questions with knowledge-based and statistical techniques [Journal Article]. Computational Linguistics. 2007;33(1):63–103.
- [7] Huang X, Lin J, Demner-Fushman D. Evaluation of PICO as a knowledge representation for clinical questions. AMIA Annual Symposium proceedings AMIA Symposium. 2006:359–63. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17238363http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC1839740.
- [8] Jin D, Szolovits P. Pico element detection in medical text via long short-term memory neural networks. In: Proceedings of the BioNLP 2018 workshop; 2018. p. 67–75.
- [9] Kim SN, Martinez D, Cavedon L, Yencken L. Automatic classification of sentences to support evidence based medicine. In: BMC bioinformatics. vol. 12. BioMed Central; 2011. p. 1–10.
- [10] Abaho M, Bollegala D, Williamson P, Dodd S. Correcting crowdsourced annotations to improve detection of outcome types in evidence based medicine. In: CEUR Workshop Proceedings. vol. 2429; 2019. p. 1–5.
- [11] Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–1240.
- [12] Beltagy I, Lo K, Cohan A. SciBERT: A pretrained language model for scientific text. arXiv preprint arXiv:190310676. 2019.
- [13] Alsentzer E, Murphy JR, Boag W, Weng WH, Jin D, Naumann T, et al. Publicly available clinical BERT embeddings [Journal Article]. arXiv preprint arXiv:190403323. 2019.
- [14] Stubbs A, Kotfila C, Uzuner O. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1 [Journal Article]. Journal of biomedical informatics. 2015;58:S11–S19.
- [15] Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification [Journal Article]. Journal of the American Medical Informatics Association. 2007;14(5):550–563.
- [16] Li J, Sun Y, Johnson RJ, Sciaky D, Wei CH, Leaman R, et al. BioCreative V CDR task corpus: a resource for chemical disease relation extraction [Journal Article]. Database. 2016;2016.
- [17] Biggane AM, Brading L, Ravaud P, Young B, Williamson PR. Survey indicated that core outcome set development is increasingly including patients, being conducted internationally and using Delphi surveys [Journal Article]. Trials. 2018;19(1):1–6.
- [18] Mascio A, Kraljevic Z, Bean D, Dobson R, Stewart R, Bendayan R, et al. Comparative Analysis of Text Classification Approaches in Electronic Health Records [Journal Article]. arXiv preprint arXiv:200506624. 2020.
- [19] Leaman R, Wei CH, Lu Z. tmChem: a high performance approach for chemical named entity recognition and normalization [Journal Article]. Journal of cheminformatics. 2015;7(S1):S3.
- [20] Wallace BC, Kuiper J, Sharma A, Zhu M, Marshall IJ. Extracting PICO sentences from clinical trial reports using supervised distant supervision [Journal Article]. The Journal of Machine Learning Research. 2016;17(1):4572–4596.
- [21] Kiritchenko S, De Bruijn B, Carini S, Martin J, Sim I. ExaCT: automatic extraction of clinical trial characteristics from journal publications [Journal Article]. BMC medical informatics and decision making. 2010;10(1):56.
- [22] Demner-Fushman D, Few B, Hauser SE, Thoma G. Automatically identifying health outcome information in MEDLINE records [Journal Article]. Journal of the American Medical Informatics Association. 2006;13(1):52–60.
- [23] Kang T, Zou S, Weng C. Pretraining to Recognize PICO Elements from Randomized Controlled Trial Literature [Journal Article]. Studies in health technology and informatics. 2019;264:188.
- [24] Brockmeier AJ, Ju M, Przybyła P, Ananiadou S. Improving reference prioritisation with PICO recognition. BMC medical informatics and decision making. 2019;19(1):1–14.
- [25] Sun C, Yang Z. Transfer learning in biomedical named entity recognition: An evaluation of BERT in the PharmaCoNER task. In: Proceedings of The 5th Workshop on BioNLP Open Shared Tasks; 2019. p. 100–104.
- [26] Melamud O, Goldberger J, Dagan I. context2vec: Learning generic context embedding with bidirectional lstm. In: Proceedings of the 20th SIGNLL conference on computational natural language learning; 2016. p. 51–61.
- [27] Peters ME, Neumann M, Iyyer M, Gardner M, Clark C, Lee K, et al. Deep contextualized word representations. arXiv preprint arXiv:180205365. 2018.
- [28] Devlin J, Chang MW, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:181004805. 2018.
- [29] Mikolov T, Chen K, Corrado G, Dean J. Efficient estimation of word representations in vector space. arXiv preprint arXiv:13013781. 2013.
- [30] Pennington J, Socher R, Manning CD. Glove: Global vectors for word representation. In: Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP); 2014. p. 1532–1543.
- [31] Doğan RI, Leaman R, Lu Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. Journal of biomedical informatics. 2014;47:1–10.
- [32] Kringelum J, Kjaerulff SK, Brunak S, Lund O, Oprea TI, Taboureau O. ChemProt-3.0: a global chemical biology diseases mapping. Database. 2016;2016.
- [33] Jin Q, Dhingra B, Cohen WW, Lu X. Probing biomedical embeddings from language models. arXiv preprint arXiv:190402181. 2019.
- [34] Sang EF, Veenstra J. Representing text chunks. arXiv preprint cs/9907006. 1999.
- [35] Williamson PR, Altman DG, Bagley H, Barnes KL, Blazeby JM, Brookes ST, et al. The COMET handbook: version 1.0 [Journal Article]. Trials. 2017;18(3):280.
- [36] Sayers E. The E-utilities in-depth: parameters, syntax and more [Journal Article]. Entrez Programming Utilities Help [Internet]. 2009.
- [37] Williamson PR, Altman DG, Blazeby JM, Clarke M, Devane D, Gargon E, et al. Developing core outcome sets for clinical trials: issues to consider. Trials. 2012;13(1):1–8.
- [38] Kirkham JJ, Dwan KM, Altman DG, Gamble C, Dodd S, Smyth R, et al. The impact of outcome reporting bias in randomised controlled trials on a cohort of systematic reviews. Bmj. 2010;340.
- [39] Dwan K, Gamble C, Williamson PR, Kirkham JJ. Systematic review of the empirical evidence of study publication bias and outcome reporting bias—an updated review. PloS one. 2013;8(7):e66844.
- [40] Stenetorp P, Pyysalo S, Topić G, Ohta T, Ananiadou S, Tsujii J. BRAT: a web-based tool for NLP-assisted text annotation. In: Proceedings of the Demonstrations at the 13th Conference of the European Chapter of the Association for Computational Linguistics; 2012. p. 102–107.
- [41] Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. arXiv preprint arXiv:170603762. 2017.
- [42] Akbik A, Blythe D, Vollgraf R. Contextual string embeddings for sequence labeling. In: Proceedings of the 27th international conference on computational linguistics; 2018. p. 1638–1649.
- [43] Ammar W, Groeneveld D, Bhagavatula C, Beltagy I, Crawford M, Downey D, et al. Construction of the literature graph in semantic scholar [Journal Article]. arXiv preprint arXiv:180502262. 2018.
- [44] Johnson AE, Pollard TJ, Shen L, Li-Wei HL, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database [Journal Article]. Scientific data. 2016;3(1):1–9.
- [45] Sharma S, Daniel Jr R. BioFLAIR: Pretrained pooled contextualized embeddings for biomedical sequence labeling tasks. arXiv preprint arXiv:190805760. 2019.
- [46] Howard J, Ruder S. Universal language model fine-tuning for text classification. arXiv preprint arXiv:180106146. 2018.
- [47] Peters ME, Ruder S, Smith NA. To tune or not to tune? adapting pretrained representations to diverse tasks. arXiv preprint arXiv:190305987. 2019.
- [48] Liu L, Mu F, Li P, Mu X, Tang J, Ai X, et al. Neuralclassifier: An open-source neural hierarchical multi-label text classification toolkit. In: Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics: System Demonstrations; 2019. p. 87–92.
- [49] Lafferty J, McCallum A, Pereira FC. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In: Proceedings of the 18th International Conference on Machine Learning (ICML 2001); 2001. p. 282–289.
- [50] Raunak V, Gupta V, Metze F. Effective dimensionality reduction for word embeddings. In: Proceedings of the 4th Workshop on Representation Learning for NLP (RepL4NLP-2019); 2019. p. 235–243.
- [51] Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. Pytorch: An imperative style, high-performance deep learning library. arXiv preprint arXiv:191201703. 2019.
- [52] van Aken B, Papaioannou JM, Mayrdorfer M, Budde K, Gers FA, Löser A. Clinical Outcome Prediction from Admission Notes using Self-Supervised Knowledge Integration. arXiv preprint arXiv:210204110. 2021.
- [53] Manning CD, Surdeanu M, Bauer J, Finkel JR, Bethard S, McClosky D. The Stanford CoreNLP natural language processing toolkit. In: Proceedings of 52nd annual meeting of the association for computational linguistics: system demonstrations; 2014. p. 55–60.
- [54] Tsuruoka Y, Tateishi Y, Kim JD, Ohta T, McNaught J, Ananiadou S, et al. Developing a robust part-of-speech tagger for biomedical text. In: Panhellenic Conference on Informatics. Springer; 2005. p. 382–392.
- [55] Kim JD, Ohta T, Tateisi Y, Tsujii J. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(suppl_1):i180–i182.
- [56] Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics. 2004;20(14):2320–2321.
- [57] Wang YX, Ramanan D, Hebert M. Learning to model the tail. In: Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017. p. 7032–7042.
- [58] Cui Y, Jia M, Lin TY, Song Y, Belongie S. Class-balanced loss based on effective number of samples. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2019. p. 9268–9277.
- [59] Lin TY, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. In: Proceedings of the IEEE international conference on computer vision; 2017. p. 2980–2988.
Appendix
Appendix A Adapting CLMs to Outcome Detection Task
1 Fine-tuning
The biomedical CLMs presented under section 3.2 are fine-tuned for the Outcome Detection (ODP) task. Given an input sentence containing words/tokens, e.g. , the CLMS are used to encode each a word to obtain a hidden state representation , where , {BERT-variants, BioELMo, BioFLAIR} and (i.e. is a vector of size ). We then apply softmax function to return a probability of each label for each position in the sentence , , where i.e. W is a matrix with dimensions (size of label set) (hidden-state size). represents the set of outcome type target labels. Given the probability distribution the softmax generates at each position, we use to to return the predicted outcome type label.
2 Building an Outcome Detection Model (ODP-tagger)
In this work, we augment a BiLSTM model with in-domain resources including medically oriented part-of-speech tags (POS) and PubMed word2vec vectors [?]. We then train the model on EBM-NLP incorporating a class distribution balancing factor which essentially aims to regularize the multiway softmax loss with a balanced weighting across multiple classes. The conscious effort of augmenting a regular BiLSTM was indeed re-enumerated with a visible gradual improvement in dev set F1 scores for the ODP task as table 10 presents. Below sections cover the augmentation steps.
Custom trained biomedical POS
We compare the performance of 3 Part-Of-Speech (POS) taggers, which include, 2 popular generic and fully established Natural language Processing (NLP) libraries, spaCy555https://spacy.io/ [?], Stanford Core NLP666https://nlp.stanford.edu/software/tagger.html [?], and a tagger specifically tuned for POS tagging tasks on biomedical text (Genia-Tagger) [?]. The Genia-Tagger is pre-trained on a collection of articles extracted from the MEDLINE database [?]. To avoid any biased analysis in the comparative study, spaCy and Stanford Core NLP are also customised for biomedical text by training them on a corpus of 6,700 Medline sentences (MedPOST) annotated with 60 POS tags [?]. These 3 taggers are each used to provide POS features to input samples (words) for a task to classify outcome phrases into five outcome types that include Physical, Pain, Mental, Mortality, Adverse effects and Other as predefined in EBM-NLP dataset. A BiLSTM network and a softmax classification layer are used to complete this task. The model using trained Stanford tagger outperforms the other two models (table 8), and as a result, we use Stanford Core NLP for POS tagging in the proceeding ODP task.
| EBM-NLP (F1%) | |
|---|---|
| BiLSTM-spaCY-MedPOST | 80.5 |
| BiLSTM-stanford-MedPOST | 81.3 |
| BiLSTM-Genia-Tagger | 79.0 |
Context-Independent PubMed word2vec vectors (W2V)
We train word2vec (W2V) on 5.5B tokens of PubMed and PMC abstracts to obtain these vectors. These fixed vectors are later replaced by the pre-trained CLMs in the feature extraction approach during evaluation.
Probing for a loss function for the ODP-tagger
We assess 3 cost-sensitive functions premised on a log-likelihood objective , (log probability of label given input word ) to identify a suitable learning loss for the ODP-tagger experiments.
| (4) |
where is the training set containing sentences, and .
Imputed Inverse loss (IIL) function
Empirically setting each labels’ weights to be inversely proportional to the label frequency. A relatively simple heuristic that has been widely adopted [?].
| (5) |
We check two variants of the scaling factor in the Imputed Inverse Loss equation , and a smoothed version , , where is the number of training samples labelled or frequency of ground truth label .
Class balanced loss (CB)
The Class balanced loss proposed by Cui et al., [?] discusses the concept of effective number of samples to capture the diminishing marginal benefits of incrementing the samples of a class. Due to the intrinsic similarities among real-world data, increasing the sample size of a class might not necessarily improve model-performance. Cui et al., [?] introduces a weighting factor that is inversely proportional to the effective number samples .
Where , , is dataset size and is the sample size of label , .
| (6) |
Focal loss (FL)
Focal loss assigns higher weights to harder examples and lower ones to the easier examples [?]. It introduces a scaling factor . is a focusing parameter in the loss function which decays to zero as the confidence in the correct class increases hence automatically down weighting the contribution of easy examples in the training and rapidly focusing on harder examples.
| (7) |
where is a weighting factor, , is set to , is the number of training samples for class , is the probability of ground truth label . We do not hypertune the focusing parameter , and instead set it to based on having achieved good results in examples [?].
| EBM-NLP | |
|---|---|
| BiLSTM | 27.0 |
| BiLSTM + | 37.0 |
| BiLSTM + | 38.0 |
| BiLSTM + CB | 37.0 |
| BiLSTM + FL | 19.0 |
Results in table 9 indicate both variants and CB are quite competitive, however we chose particularly because it slightly outperforms all the other tested for the objective loss function.
Introducing an undersampling hyper-parameter (US)
In this strategy, we randomly undersample the majority class of the dataset by a specified percentage. The objective of the ODP-tagger is to minimize the Imputed Inverse loss (IIL2) derived from the preceding section which probes for a suitable loss function,
| (8) |
| Model | F1 | |
| 1 | BiLSTM | 32.5 |
| 2 | BiLSTM + POS | 37.9 |
| 3 | BiLSTM + POS + W2V | 41.1 |
| 4 | BiLSTM + POS + W2V + | 43.2 |
| 5 | BiLSTM + POS + W2V + + | 43.6 |
| 6 | BiLSTM + POS + W2V + + + CRF | 44.0 |
| 7 | BiLSTM + POSSt + W2VPb + | 42.8 (1.5) |
| 8 | BiLSTM + POSSt + W2VPb + + | 43.2 (1.9) |
| 9 | BiLSTM + POSSt + W2VPb + + + CRF | 44.3 (1.4) |
Table 10 results are emblematic of the positive impact each of the different strategies had in architecting the ODP-tagger. We observe slight performance improvements upon adopting (a strategy in which the majority class is undersampled by 50% during training) and replacement of the softmax with a CRF for classification. We observe cumulative gains in performance of 5.4%, 3.2% and 2.1% upon adding POSSt, W2VPb and respectively. On the otherhand, adopting and replacement of the softmax with a CRF for classification lead to slight improvements of 0.4% each.
We are aware that the improvements narrated above can dramatically change given new splits of the data, particularly the slight improvements brought about by and the CRF. Therefore, to account for this, we check for the robustness of the improvements brought about by and the CRF by measuring performance across 5 different randomly split train and test sets. The mean and (standard deviation) across the 5 experiments of the random splits are reported in Exps 7, 8 and 9. Results obtained in 8 and 9 show that both and the CRF respectively lead to substantial improvements in performance when added to the ODP-tagger. Later on, we hypertune multiple parameters to obtain the optimal parameter settings (11) for fine-tuning and feature extraction experiments.
Appendix B Hyper-parameter Tuning
The tuned ranges for the hyper-parameters used in our models are included in table 11.
| Fine-tuning | ||
| Tuned range | Optimal | |
| Learning rate | [1e-5,1e-4, 1e-3, 1e-2] | 1e-5 |
| Train Batch size | [16, 32] | 32 |
| Epochs | [3, 5, 10] | 10 |
| Sampling % (US) | [50, 75, 100] | 100 |
| Optimizer | [Adam, SGD] | Adam |
| ODP-tagger | ||
| Learning rate | [1e-4, 1e-3, 1e-2, 1e-1] | 1e-1 |
| Train Batch size | [50, 150, 250, 300] | 300 |
| Epochs | [60, 80, 120, 150] | 60 |
| Sampling % (US) | [10, 25, 50, 75] | 50 |
| Optimizer | [Adam, SGD] | SGD |
Appendix C A classification taxonomy of outcome domains suitable for retrieval of outcome phrases from clinical text
| Core area | Outcome domain | Domain symbol | Explanation | |||||||||||
| Physiological | Physiological/Clinical | P 0 |
|
|||||||||||
| Death | Mortality/survival | P 1 |
|
|||||||||||
| Life impact | Physical functioning | P 25 |
|
|||||||||||
| Social functioning | P 26 |
|
||||||||||||
| Role functioning | P 27 |
|
||||||||||||
| Emotional functioning/wellbeing | P 28 |
|
||||||||||||
| Cognitive functioning | P 29 |
|
||||||||||||
| Global quality of life | P 30 |
|
||||||||||||
| Perceived health status | P 31 |
|
||||||||||||
|
||||||||||||||
| Personal circumstances | P 33 |
|
||||||||||||
| Resource use | Economic | P 34 |
|
|||||||||||
| Hospital | P 35 |
|
||||||||||||
| Need for further intervention | P 36 |
|
||||||||||||
| Societal/carer burden | P 37 |
|
||||||||||||
| Adverse events | Adverse events/effects | P 38 |
|