Adaptive and Efficient Learning with Blockwise Missing and Semi-Supervised Data
Abstract
Data fusion is an important way to realize powerful and generalizable analyses across multiple sources. However, different capability of data collection across the sources has become a prominent issue in practice. This could result in the blockwise missingness (BM) of covariates troublesome for integration. Meanwhile, the high cost of obtaining gold-standard labels can cause the missingness of response on a large proportion of samples, known as the semi-supervised (SS) problem. In this paper, we consider a challenging scenario confronting both the BM and SS issues, and propose a novel Data-adaptive projecting Estimation approach for data FUsion in the SEmi-supervised setting (DEFUSE). Starting with a complete-data-only estimator, it involves two successive projection steps to reduce its variance without incurring bias. Compared to existing approaches, DEFUSE achieves a two-fold improvement. First, it leverages the BM labeled sample more efficiently through a novel data-adaptive projection approach robust to model misspecification on the missing covariates, leading to better variance reduction. Second, our method further incorporates the large unlabeled sample to enhance the estimation efficiency through imputation and projection. Compared to the previous SS setting with complete covariates, our work reveals a more essential role of the unlabeled sample in the BM setting. These advantages are justified in asymptotic and simulation studies. We also apply DEFUSE for the risk modeling and inference of heart diseases with the MIMIC-III electronic medical record (EMR) data.
Keywords: Missing Data; Data fusion; Control variate; Calibration; Linear allocation; Statistical Efficiency.
1 Introduction
1.1 Background
Data fusion enables more powerful and comprehensive analysis by integrating complementary views provided by different data sources, and has been more and more frequently adopted with the increasing capacity in unifying and synthesizing data models across different cohorts or institutions. As an important example, in the field of health and medicine, there has been growing interests and efforts in linking electronic medical records (EMRs) with biobank data to tackle complex health issues (Bycroft et al.,, 2018; Castro et al.,, 2022, e.g.). EMRs include detailed longitudinal clinical observations of patients, offering a rich source of health information over time. By combining these records with the genetics, genomics and other multi-omics data in biobanks, this type of data fusion enables a much deeper understanding of disease prognosis at the molecular level, potentially transforming patient care and treatment strategies (Zengini et al.,, 2018; Verma et al.,, 2023; Wang et al.,, 2024, e.g.). In addition to combine different types of data, data fusion has also been scaled up to merge data from multiple institutions. The UK Biobank (Sudlow et al.,, 2015) and the All-of-Us Research Program (The All of Us Research Program Investigators,, 2019) in the U.S. are prime examples.
To robustly and efficiently learn from multi-source fused data sets, several main statistical challenges need to be addressed. One is blockwise missing (BM) often occurring when certain variables are collected or defined differently across the sources. Consequently, one or multiple entire blocks of covariates are missing in the merged dataset. Dealing with the paucity of accurate outcomes in EMR is another major problem. This is due to the intensive human efforts or time costs demanded for collecting accurate outcomes like the manual chart reviewing labels created by experts for certain diseases or a long-term endpoint taking years to follow on. In this situation, one is often interested in semi-supervised (SS) learning that combines some small labeled subset with the large unlabeled sample collected from the same population without observations of the true outcome. In the aforementioned EMR applications, it is often to have the BM and SS problems occurring together, e.g., our real example in Section 5. This new setup has a more complicated data structure to be formally described in Section 1.2 and motivates our methodological research in this paper.
1.2 Problem Setup
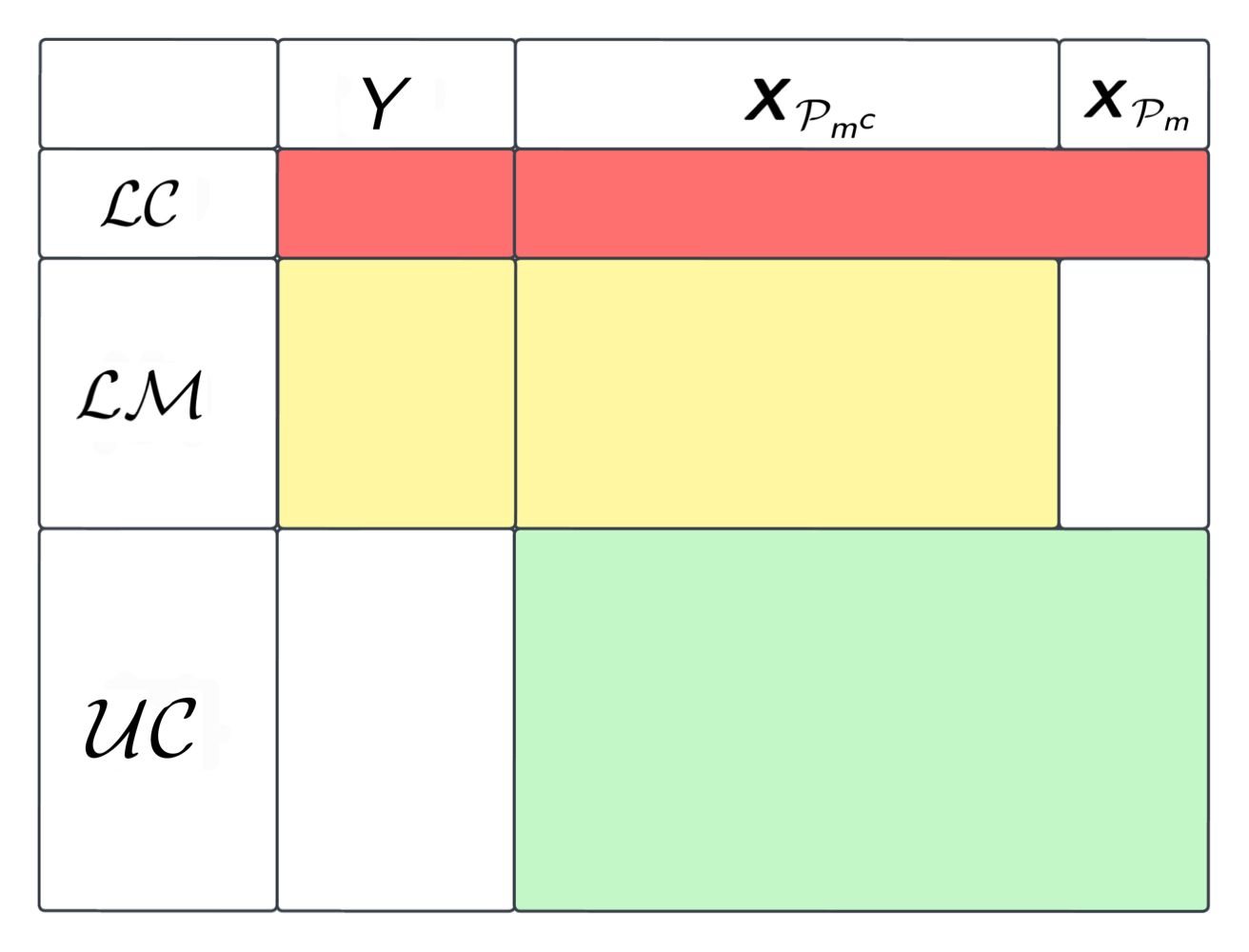
Let be the outcome of interest and be a vector of -dimensional covariates. As illustrated on the left panel of Figure 1, there are three data structures of the observations from the same population: (i) observations of that are labeled and complete, denoted as ; (ii) labeled and covariates-missing observations of denoted as where is the index set of the blockwise missing covariates and ; and (iii) unlabeled sample with complete observation of the covariates denoted as . Note that we consider a missing completely at random (MCAR) regime with the , and samples having a homogeneous underlying distribution. In the SS setting, we assume that . Denote by . In our application, and have similar magnitudes while our method and theory also accommodate the and scenarios. Let , and the observed samples as
where indicates the data type of the observation . Also, we are interested the scenario with multiple BM structures: with each representing a subset of data with observations of and missing covariates on the index set satisfying for any pair of in ; see the right panel of Figure 1.

Our primary interest lies in estimating and inferring the generalized linear model (GLM):
| (1) |
where is a vector of unknown coefficients and is a known link function. For example, corresponds to the Gaussian linear model and for the logistic model. It is important to note that our framework allows the model (1) to be misspecified, and we actually define the model coefficients of our interests as , the solution to the population-level moment equation:
| (2) |
The model parameter can be identified and estimated using the standard regression on the sample but and cannot be directly incorporated into this procedure, which results in loss of effective samples. In this paper, our aim is to leverage both the and samples to enhance the estimation efficiency of without incurring bias. As will be introduced in Section 5.1, the data in our setup can also include auxiliary features that are predictive of and , observed on all subjects, and possibly high-dimensional. These variables are not included in the outcome model of our primary interest due to scientific reasons but can be simply incorporated into our framework to enhance estimation efficiency. For the ease of presentation, we drop the notation of such auxiliary features in the main body of our method and theory.
1.3 Related Literature
With the increasing ability in collecting data with common models across multiple institutions, integrative regression with blockwise missing covariates has been an important problem frequently studied in recent literature. One of the most well known and commonly used approach to address missing data is Multiple Imputation with Chained Equations (MICE) White et al., (2011). However, as a practical, general, and principal strategy, MICE is sometimes subjected to high computation costs as well as potential bias and inefficiency caused by the misspecification or excessive error in imputation, e.g., see our simulation results.
Recently, Yu et al., (2020) proposed an approach named DISCOM for integrative linear regression under BM. It approximates the covariance of through the optimal linear combination across different data structures and incorporates it with an extended sparse regression to derive an integrative estimator. Xue and Qu, (2021) proposed a multiple blockwise imputation (MBI) approach that aggregates linear estimating equations with multi-source imputed data sets constructed according to their specific BM structures. They justified the efficiency gain of their proposal over the complete-data-only estimator. Xue et al., (2021) extended this framework work for high-dimensional inference with BM data sets. Inspired by the idea of debiasing, Song et al., (2024) developed an approach for a similar problem without imputing the missing blocks but showing better efficiency. Jin and Rothenhäusler, (2023) proposed a modular regression approach accommodating BM covariates under the conditional independence assumption across the blocks of covariates given the response.
All works reviewed above primarily focused on the linear model, which is subtly different from the GLM with a nonlinear link as the latter typically requires the sample to ensure identifiability of the outcome model while the linear model does not. For integrative GLM regression with BM data, Li et al., 2023b proposed a penalized estimating equation approach with multiple imputation (PEE–MI) that incorporates the correlation of multiple imputed observations into the objective function and realizes effective variable selection. Kundu and Chatterjee, (2023) and Zhao et al., (2023) proposed to incorporate the sample through the generalized methods of moments (GMM) that actually uses the score functions of a reduced GLM (i.e., the parametric GLM for ) on the sample for variance reduction. Such a “reduced model” strategy has been used frequently in other work like Song et al., (2024), as well as for fusing clinical and observational data to enhance the efficiency of causal inference (Yang and Ding,, 2019; Hatt et al.,, 2022; Shi et al.,, 2023, e.g.).
As will be detailed in our asymptotic and numerical studies, the above-introduced reduced model or imputation strategies does not characterize the true influence of the data and, thus, not achieving semiparametric efficiency in the BM regression setting, which dates back to the seminal work by Robins et al., (1994) and has been explored (Chen et al.,, 2008; Li and Luedtke,, 2023; Li et al., 2023a, ; Qiu et al.,, 2023, e.g.) in various settings. In particular, Li and Luedtke, (2023) proposed derived the canonical gradient for a general data fusion problem and justified its efficient. Nevertheless, such semiparametric efficiency property is established under an ideal scenario with a correct conditional model for , which is stringent in practice. Their method could be less robust and effective in a more realistic case when the imputation for is not consistent with the truth. This issue has been recognized and studied in recent literature mainly focusing on SS inference (Gronsbell et al.,, 2022; Schmutz et al.,, 2022; Song et al.,, 2023; Angelopoulos et al.,, 2023; Miao et al.,, 2023; Gan and Liang,, 2023; Wu and Liu,, 2023). Notably, in our real-world study including high-dimensional auxiliary features to impute the missing variables (see Section 5.1), this problem is even more pronounced due to the difficulty of high-dimensional modeling. In this case, it is challenging to realize adaptive and efficient data fusion with above-mentioned strategies, which can be seen from the results in Section 5.
Our work is also closely related to recent progress in semi-supervised learning (SSL) that aims at using the large unlabeled sample to enhance the statistical efficiency of the analysis with relatively small labeled data sets. On this track, Kawakita and Kanamori, (2013) and Song et al., (2023) proposed to use some density ratio model matching the labeled and unlabeled samples to reweight the regression on the labeled sample, which, counter-intuitively, results in SSL estimators with smaller variances than their supervised learning (SL) counterparts. Chakrabortty and Cai, (2018) and Gronsbell et al., (2022) proposed to incorporate the unlabeled sample by imputing its unobserved outcome with an imputation model constructed with the labeled data. Extending this imputation framework, Cai and Guo, (2020), Zhang and Bradic, (2022) and Deng et al., (2023) studied the SSL problem with high-dimensional , and Wu and Liu, (2023) addressed the SSL of graphic models.
Though having different constructions, these recent SSL approaches achieve a very similar adaptive property that they can produce SSL estimators more efficient than SL when the outcome model for is misspecified and as efficient as SL when it is correct. Azriel et al., (2022) discussed this property and realized it for linear models in a more flexible and effective way. Notably, our SSL setting with BM designs has not been studied in this track of SSL literature. As will be shown later, this problem is essentially distinguished from existing work in SSL as the efficiency gain by using the sample is attainable even when the outcome model (1) is correctly specified in our case. In addition, we notice that Xue et al., (2021) and Song et al., (2024) also considered using large samples to assist learning of high-dimensional linear models under the BM setting. Unlike us, they only used the sample to combat and reduce the excessive regularization errors in sparse regression and debiased inference. Nevertheless, for the inference of a low-dimensional projection of the model parameters, the asymptotic variance still remains unchanged compared to the SL scenario after incorporating the sample in their methods.
1.4 Our contribution
Regarding the limitation of existing work discussed in the previous section, we propose a Data-adaptive projecting Estimation approach for data FUsion in the SEmi-supervised setting (DEFUSE). Our method contains two major novel steps, it initiates from the simple -only estimator, and first incorporates the sample to reduce its asymptotic variance through a data-adaptive projection procedure for the score functions. Then, it utilizes the large sample in another projecting step, to further improve the efficiency of the data-fused estimator resulting from the previous step. In the following, we shall summarize the main novelty and contribution of our work that have been justified and demonstrated through the asymptotic, simulation, and real-world studies.
-
1.
Our approach in combining the and samples is shown to produce an asymptotically unbiased and more efficient estimator compared to recent developments for BM regression (Xue and Qu,, 2021; Zhao et al.,, 2023; Song et al.,, 2024, e.g.). The newly proposed framework is inspired by the semiparametric theory and allows the flexible use of general ML algorithms to impute the missing variables. The efficiency gain of our method over existing methods in various settings like model misspecification are justified from both the theoretical and numerical perspectives.
-
2.
We develop a novel data-driven control function approach to realize adaptive and efficient estimation under potential misspecification or poor quality of the imputation models, especially when there exists nonlinear relationship not captured by them. Compared to recent work in addressing problematic nuisance models (Angelopoulos et al.,, 2023; Miao et al.,, 2023, e.g.), our strategy is shown to realize more flexible and efficient calibration in our theory, simulation, and real-world studies. This improvement is even clearer when high-dimensional auxiliary features are included in the nuisance model like our EMR application.
-
3.
We establish a novel framework to bring in large unlabeled samples under the SS setting and leverage them to enhance the efficiency of the BM estimation. This enables a more efficient use of the sample leading to an estimator of strictly smaller variance than that obtained only using and , which has not been readily achieved in previous work on a similar setting (Xue et al.,, 2021; Song et al.,, 2024). Our development is also shown to be more efficient than existing approaches for the standard SS setting without BM (Gronsbell et al.,, 2022, e.g.), in both theoretical and numerical studies.
-
4.
We propose a linear allocation approach to extend our framework in addressing multiple BM structures, in which the classic semiparametric theory does not apply.
2 Method
2.1 Outline and Preliminary Framework
Without the loss of generality, we aim at estimating and inferring a linear projection of the model coefficients denoted by where is an arbitrary vector satisfying . In this framework, one can take as the -th unit vector in to infer each GLM effect and can also take as the observed covariates of some new coming subject for the purpose of prediction. Let represent the empirical mean of some measurable function on the data set that can be taken as one or the union of more than one belonging to , e.g.,
Similarly, we use to represent the empirical variance of on data set , e.g., . We start from introducing the DEFUSE method for the scenario with a single data set; see the left panel of Figure 1. It includes three steps outlined in Figure 2 and also in Algorithm 1 presented later.

First, we solve the empirical version of the estimating equation (2) for based on the sample:
| (3) |
Denote the solution as . It is also the maximum likelihood estimator (MLE) for the Gaussian linear and logistic models when and respectively. Using the standard asymptotic theory for M-estimation (Van der Vaart,, 2000), we have the expansion
which will be more strictly given by Lemma 1. Consequently, the asymptotic variance of is proportional to that of the score function . Therefore, our basic idea of incorporating the sample is to leverage its observations of to form some function explaining the variance of as much as possible, and use it as a control variate to adjust for variance reduction.
In specific, we let and . Motivated by semiparametric theory (Robins et al.,, 1994, e.g.), the ideal control function in this case is (with empirically form ), with the corresponding estimator incorporating formed as
| (4) |
Here, the augmentation term can be viewed as a control variate of mean zero and negatively correlated with the empirical mean of the score on . Among all the control variate in this form, provides the optimal choice to achieve the semiparametric efficiency. However, a critical concern in this classic framework is that computing requires the knowledge of the conditional distribution . It is typically unknown in practice and needs to be replaced with some data generation model where is pre-specified and is an empirical estimate of the unknown model parameters. Correspondingly, our imputation of is formed as
| (5) |
and to be used as a control function for the variance reduction of . In the following, we remark and discuss on the constructing strategy and computation of and in practice.
Remark 1.
The data imputation model can be either a simple parametric model with finite-dimensional or more sophisticated ones with diverging parameter space. For example, for continuous , one can either (i) assume that follows a (multivariate) Gaussian linear model against and learn it through the a standard MLE; or (ii) assume with independent from , then use some ML algorithm (e.g., random forest or neural network) to learn the mean model and the standard method of moment on the fitted residuals to estimate ; or (iii) implement generative ML like conditional generative adversarial network (GAN) (Mirza and Osindero,, 2014) to learn without any model assumptions.
Moreover, as will be discussed and implemented in Section 5, can be constructed with auxiliary covariates not belonging to included in the targeted outcome model. This may invoke learning with high-dimensional features and excessive errors, which highlights the importance of carrying out flexible data-adaptive calibration to be introduced in Section 2.2. Regarding these choices, our asymptotic analysis generally accommodate all the three types with or without auxiliary covariates.
When using complex ML algorithms like random forests to learn , we adopt cross-fitting to avoid the over-fitting bias as frequently done in related contexts (Chernozhukov et al.,, 2016; Liu et al.,, 2023, e.g.). In specific, we randomly split the sample into folds with an equal size, denoted as . For each , we implement the ML method on to obtain an estimator . Then for each from , we use as its score imputation since is independent with . For each from , we use the average imputation . As shown in Section 3, this strategy is important to eliminate the over-fitting bias of arbitrary ML methods. For the ease of presentation, we slightly abuse the notation in the remaining paper by dropping the superscript of related to cross-fitting.
With , one can use either numerical methods like Gaussian quadrature or Monte-Carlo (MC) sampling to compute . When includes many covariates, the computational burden and accuracy through either way seems to be a challenging issue. However, inspecting the form of the score function , we notice that it depends on the entire only through at most two-dimensional linear representations and . Thus, both Gaussian quadrature and MC can achieve good computational performance thanks to this low dimensionality (Caflisch,, 1998; Kovvali,, 2022). For MC, we could also use the same set of MC samples on all subjects in certain scenarios (e.g., Gaussian noise under homoscedasticity) to save computational costs. In our case, we choose the MC method to compute as it facilitates our data-adaption procedure to be introduced next.
2.2 Data-adaptive Calibration
The numerical approximation of (5) offers a convenient approach for estimating the conditional expectation function . However, the nuisance model , whether parametric or nonparametric, can deviate from the truth in practice due to model misspecification or excessive ML errors. This deviation typically does not cause excessive bias (as shown in our asymptotic analysis) but could result in inefficient estimation in (4). Similar observations have been made in recent missing data literature (Angelopoulos et al.,, 2023; Wu and Liu,, 2023, e.g.). To address this issue, we propose a novel data-adaptive calibration approach to further reduce the variance in the presence of model misspecifications. The key idea is to introduce a broader class of conditional model for with additional unknown parameters , denoted as . We can then calibrate through its free parameter to produce a more efficient estimator than (4) with wrong .
In specific, let be independent random draw from the preliminary for the th subject in . The standard MC approximation of defined in (5) is given by In Section 3, we show that a size moderately increasing with the sample size is sufficient to guarantee the asymptotic normality and efficiency of our estimator. Inspired by importance weighting, we re-construct the control function with our newly introduced as follows
Its MC approximation is given by
where and is known as the importance weighting (IW) function such that
Following the definition of , we can construct as the product of and an IW function that is only parameterized by . It is easy to see that when , . Hence, encompasses a broader class of conditional models of . We then replace in (5) by , and calibrate it by optimizing as follows.
| (6) |
By design, maximize the correlation between and while minimizing the variance of itself. Consequently, the new control variable with optimized can further reduce the estimation variance when is misspecified. We define the corresponding estimator in a single BM scenario as
| (7) |
and denote it as DEFUSE1.
Just like , the IW model can also be constructed using general learners (cross-fitting needed for complex ML) as long as the constant belongs to their feasible spaces or sets. Without loss of generality, we impose this constraint as . As will be shown later, this can ensure that when is consistent with the true conditional distribution of , we have and the resulting to be asymptotically equivalent with the semiparametric efficient estimator (4). Similarly, it is also tentative to make feasible in our construction since it can ensure to have no larger variance than the -only estimator , i.e., avoiding negative effects of incorporating . For the choice of , one can see Assumption 5 and Lemma 2 for more rigorous details.
Notably, even when has negative values or an integral not equal to , can still be asymptotic unbiased and of effectively reduced variance. In general, (6) can be a non-convex problem with a high computational cost to optimize due to the MC sampling. To handle it, one may use stochastic gradient descendant (SGD) that generates a few MC samples for each observation (i.e., with small ) and makes gradient update accordingly in multiple iterations. We shall remark on practical choices on and subject to our computational concerns.
Remark 2.
To avoid the high computational costs and potential over-fitting, we recommend setting to have a simpler form than . A simple but usually effective choice is to use exponential tilting as described in (Johns,, 1988), where and is a -dimensional basis with . However, this makes (6) non-convex and hence challenging to solve. Therefore, we propose further simplifying it to , which makes (6) a convex problem with an explicit-form solution. This form closely resembles the exponential one when is small. Additionally, when include a constant term of for intercept, both and are feasible solutions.
2.3 Semi-supervised Estimation
After obtaining DEFUSE1, we further incorporate the sample to enhance its efficiency. This also involves a data-adaptive projection step that shares a similar spirit as in DEFUSE1. For the -sample terms composing in (7), an interesting observation is that the score function of is orthogonal to when the outcome model (1) is correct. This orthogonality leads to limited efficiency gains in the existing SSL methods for standard GLMs, such as (Chakrabortty and Cai,, 2018), because their estimates are score-function based. However, the projection of on , is typically not orthogonal to . Thus, unlike those existing SSL methods, can partially explain the variance of through the projection, even when the outcome model is correct. This promises the benefit of leveraging the large sample in our estimates.
Inspired by this, we propose to model the conditional distribution given , denoted as , which can be either directly specified as the outcome model (1) with some extra nuisance parameters (e.g., variance of Gaussian noise), or estimated using ML as discussed in Remark 1. Similar as in Section 2.2, we introduce an IW function
where stands for the calibrated conditional model for to improve the efficiency when is misspecified. The optimal can be obtained by
| (8) |
where is the empirical score function of DEFUSE1 in the sample. The term is constructed as where are i.i.d. random drawn from the preliminary model .
Comparing to (6), the objective function in (8) does not include the variance term of . That is because the variance of on is negligible in the SS setting with . Finally, the DEFUSE estimator in the single BM scenario is constructed as
| (9) |
Again, we impose so that can achieve semiparametric efficiency in this SS step when the imputation distribution is consistent with the truth. Also, letting constant be in the feasible space of can ensure that is feasible and has a smaller or equal variance than . For specification of , we suggest using similar strategies as proposed in Remark 2. We end our exposition for the single BM and SS setting with a remark on our constructing strategy.
Remark 3.
Our framework starts from leveraging the sample by projecting to and then incorporates to further enhance the resulting DEFUSE1 estimator. Reversing the order of these two projection steps is likely to result in a less efficient estimator because the score of tends to be orthogonal to and directly using on it may not be helpful when the outcome model (1) is strictly or approximately correct. Also, we notice that is orthogonal to when the models for and are both correct and its empirical version tends to be poorly explained by on the data in our finite sample studies. Therefore, we do not elaborate the variance contributed by the sample in our SS procedure to save computation costs.
2.4 DEFUSE in the Multiple BM Scenario
In this section, we extend the proposed DEFUSE method to the scenario of data sets with different BM sets denoted as and sample sizes ; see the right panel of Figure 1. Same as the single BM scenario, we start with the standard -only estimator and leverage ’s to improve its efficiency. To this end, we first implement data-adaptive calibration proposed in Section 2.2 with each separately, to obtain the control function analog to in the single BM case. Here we drop the notations of the nuisance parameters in for simplification.
Denote by . Then we assemble the multiple control functions to form the DEFUSE1 estimator as
| (10) |
where the ensemble weights ’s in are solved from the quadratic programming:
| (11) |
and . The goal of (11) is to find the optimal allocation minimizing the asymptotic variance of DEFUSE1 as formed in (10) that augments the preliminary with a linear combination of the variance reduction term coming from every . Correspondingly, the objective function in (11) is set as the empirical variance of such an estimator with weights ; see our asymptotic analysis for details.
One can view (11) as a ridge regression with each coefficient penalized according to the sample size and variance of . For certain BM patterns, can have highly correlated or collinear elements. In this case, the “ridge penalty” on can actually ensure the non-singularity and stability of (11). Thus, we assign an upper bound to each in our definition so that cannot be too small to make this penalty ineffective even when .
Similarly to the single BM scenario, we incorporate the sample to further reduce the variance of using the same idea in Section 2.3. Note that the empirical score function of on can be written as . Naturally, we use it to replace the score in (8) and solve it to obtain corresponding calibration parameters. Then we use this solution to impute the score and augment in the same form as (9). This procedure can be regarded as a straightforward extension of our proposal in Section 2.3.
We summarize the main steps of the DEFUSE approach in Algorithm 1. We notice that some recent work like Kundu and Chatterjee, (2023) highlighted practical needs to protect the privacy of individual-level data when fusing the data sets coming from different data institutions. This has been achieved by transferring summary data only (e.g., mean vectors, model coefficients, and covariance matrices) across the sites, which is often referred as the DataSHIELD framework or constraint Wolfson et al., (2010). Our method can naturally accommodate this situation as well. In specific, note that Steps 1, 2.1, and 3.1 in Algorithm 1 only rely on the sample while Steps 2.2 and 3.2 require summary data (i.e., the empirical mean of the imputed score functions) from each or . In practice, and are likely to be stored in the same site. Thus, under the data-sharing constraint, the implementation of DEFUSE requires usually one and at most two rounds of communication between and other sites, which is communication efficient and user-friendly.
3 Asymptotic Analysis
3.1 Notations and Assumptions
In this section, we conduct theoretical analysis on the asymptotic normality and efficiency gain of DEFUSE. Justification of all the results presented in this section can be found in Appendix. We begin with notations and main assumptions. Let denote the true probability density or mass function of random variables and be the conditional probability function, be the population covariance operator, and be the diagonal matrix with its -th diagonal entry being . For simplicity of presentation, denote by . For two sequences and with index , we say when there exists some constant such that , when , for , and or for or with the probability approaching . We say is a Lipschitz function if there exists a constant such that for any and in the domain of , .
We first introduce the assumptions for analyzing DEFUSE in the single BM scenario. Assume that converges to some , , and the number of MC sample with an arbitrary constant that can be set small to save computational costs without having impacts on the asymptotic properties of DEFUSE.
Assumption 1.
The derivative of the link function is Lipschitz; belongs to a compact space including as its interior; and the Hessian has all its eigenvalues staying away from and . The joint distribution of has a continuously differentiable density for the continuous components and satisfies that
Assumption 2.
There exists some population level parameters and such that
where we denote by
Assumption 3.
There exists some population level parameters and such that
where and .
Remark 4.
Assumption 1 include mild and common regularity conditions for M-estimation (Van der Vaart,, 2000). Assumptions 2 and 3 convey a message that to establish the asymptotic normality of DEFUSE, all the nuisance estimators used for imputation and calibration need to be consistent with some limiting values. Compared to the double machine learning (DML) framework (Chernozhukov et al.,, 2018), we consider a more stringent MCAR regime, under which the estimation forms (7) and (9) automatically correcting the excessive bias of the nuisance estimators (with cross-fitting for general ML). Consequently, our assumption on the quality of the nuisance estimators is substantially weaker than DML from two aspects. First, we only require them to converge to some population limits but not necessarily the true distributions required by DML. Second, we assume an -convergence rate essentially slower than the rate assumed in DML.
Remark 5.
Assumption 2 on the consistency of the preliminary imputation models is reasonable and justifiable across broad settings with and constructed using either classic regression or modern ML tools. One can refer to comprehensive literature for the consistency of various learning approaches under potentially misspecification, e.g., parametric GLMs (Tian et al.,, 2007, e.g.); kernel methods (Zhang et al.,, 2023, e.g.); high-dimensional sparse regression (Negahban et al.,, 2012, e.g.); and random forest (Athey et al.,, 2019, e.g.).
For the calibration estimators and , we present Proposition 1 to justify their consistency in Assumption 3 under a parametric form satisfying mild regularity conditions.
Proposition 1.
Based on the population-level parameters introduced in Assumptions 2 and 3, we further define the population control and score functions as
| (12) |
which will be used later to characterize the asymptotic properties of DEFUSE. Finally, we introduce an additional Assumption 4 used for asymptotic analysis of the multiple BM scenario in Section 2.4.
Assumption 4.
In the multiple BM setting, it is satisfied: (i) Our data and construction for all BM components in satisfy Assumptions 1–3 for in the single BM scenario; (ii) Each converges to some for ; (iii) Given that (i) and (ii) hold, define as the limiting function for each analog to in (12), and denote by . Then matrix
has all its eigenvalues staying away from .
3.2 Asymptotic Normality
In this section, we present the asymptotic properties of the proposed estimators, with their efficiency analyzed and articulated in the next section. We start from the standard M-estimation result of the -only estimator in Lemma 1 that is already well-established (Van der Vaart,, 2000, e.g.).
Next, we establish the consistency and asymptotic normality for the DEFUSE1 and DEFUSE estimators in the single BM scenario.
Theorem 1 states that our DEFUSE estimators are -consistent, asymptotically unbiased and normal no matter the imputation distributions are correct or not and even when the nuisance estimators have slow convergence rates. The efficiency of and will be analyzed and discussed based on Theorem 1 later. As an extension, we also establish an asymptotic theorem for in the multiple BM setting. Its downstream SS estimation can be justified in basically the same way as the analysis on in Theorem 1 so we omit this part to avoid redundancy.
Theorem 2.
3.3 Relative Efficiency
Now we shall demonstrate the relative efficiency of the proposed DEFUSE estimators compared to a broad set of existing ones including: (a) the -only estimator ; (b) the standard semiparametric efficient estimator (Robins et al.,, 1994) based on (4) with simply imputed by ; (c) BM estimators constructed in different strategies (Xue and Qu,, 2021; Zhao et al.,, 2023), as well as Xue et al., (2021); Song et al., (2024) also considering the SS setting; (d) SS methods only incorporating the sample (Gronsbell et al.,, 2022, e.g.). In addition, we will remark on the connection of our data-adaptive calibration strategy to some recent work for the SS inference. We begin with the assumptions and lemma on the feasible set and regularity of the calibration models.
Assumption 5.
The population-level parameters and defined in Assumption 2 are the unique minimizer of the variance functions and respectively, where
Assumption 6.
Constant belongs to the feasible sets of the calibration models and we can set and without loss of generality. If some belongs to the feasible set of or , then the function also belongs to that feasible set for any .
Lemma 2.
Though imposing linear specification on and , Lemma 2 can be generalized to other forms like but this may encounter local minima issues in practice. The regularity assumptions on and in Lemma 2, i.e., Assumption A1 is mild and tends to hold under proper choices.
Remark 6.
We notice that some recent approaches for power enhancement under misspecified nuisance models leverage similar properties as Assumptions 5 and 6. For example, Gronsbell et al., (2022) combined the supervised and SS estimators through their optimal convex allocation with minimum variance. We can show that their strategy is essentially similar to setting our with only one parameter to fit. This easily satisfies above-introduced assumptions on (with in Assumption 6). As a result, our following corollaries about efficiency may apply in their case. Similar constructions could be found also in Miao et al., (2023) and others. Consequently, when taking as a nested model of the most simple one , e.g., , one could get more efficient estimators through our strategy. That means, our work actually implies a more flexible and powerful framework for nuisance model calibration in a broader set of missing data problems like SS inference.
Now we shall introduce several corollaries of Section 3.2 to characterize the efficiency of the DEFUSE estimators. We define as the relative efficiency between two asymptotically normal estimators and , where represents their asymptotic variances. First, we focus on the single BM scenario and compare the asymptotic variances of the DEFUSE1 and -only estimators in Corollary 1. We show that their is ensured to be larger than and increase along with the ratio . This is a natural result as will reduce our construction of DEFUSE1 to the -only and utilizing more samples can amplify our efficiency gain over not depending on .
Corollary 1 (DEFUSE1 v.s. -only).
Then, still in the single BM scenario, we connect DEFUSE1 with the classic semiparametric efficient estimator obtained by replacing the truth with its estimate in (4). We show that DEFUSE1 will be as efficient as in the ideal case that is consistent with . More importantly, is guaranteed to be no worse and typically more efficient than when the conditional model for is wrongly specified or erroneous and does not converge to the truth. Similar efficiency properties can be justified for our step of leveraging to construct ; see Appendix.
Corollary 2.
Remark 7.
By Corollary 2, DEFUSE1 reaches the semiparametric efficiency bound (Robins et al.,, 1994) when is consistent with the truth. This desirable property was not readily achieved by recent BM fusion approaches relying on other fusing strategies (Xue and Qu,, 2021; Zhao et al.,, 2023; Song et al.,, 2024, e.g.). This implies that with correct nuisance models, is ensured to be smaller those of the existing methods. Our numerical results in next sections support this point. Nevertheless, our method requires the observation of samples while methods like Xue and Qu, (2021) consider a more general case not necessarily having the data. Also, our method tends to have higher computational costs due to MC sampling.
In Corollary 3, we quantify the efficiency gain of incorporating the sample to further enhance . Based on this, all above corollaries and remarks on will naturally hold for our final DEFUSE estimator . We also present the asymptotic variance of in the ideal case that both the conditional models and are consistent with the true distributions.
Corollary 3.
Remark 8.
Recent work including Xue et al., (2021) and Song et al., (2024) also considered the SS and BM setting and only leveraged the large sample to assist debiased inference of high-dimensional linear models. The efficiency gain of the SS estimator over stated in Corollary 3 has not been readily achieved in earlier work on this track. For the standard SS setting, methods proposed by Chakrabortty and Cai, (2018) and others have achieved the semiparametric efficiency bound when is correct. We show in Corollary 3 that when both and are consistent with the truths. Meanwhile, we demonstrate in our simulation and real-world studies that such an efficiency gain of DEFUSE over existing SS approaches (Gronsbell et al.,, 2022; Miao et al.,, 2023, e.g.) is generally significant, no matter the nuisance models are correct or not.
At last, we introduce the efficiency property of in the multiple BM setting. In Corollary 4, we establish the optimality of the linear allocation weights for ’s, and show that is more efficient than the -only . Although the classic semiparametric theory is only applied to the single BM scenario (see Section 6 for more discussion), we still find in numerical studies that our way of constructing the control functions for each as well as the linear allocation strategy on can help to improve the performance over existing multiple BM methods not relying on the semiparametric construction; see Section 4.2 for details.
Corollary 4.
Under Assumption 4 and that each , achieves the smallest asymptotic variance among the linear allocation estimators formed as
Also, , where holds when at least one among have non-zero correlations with .
4 Simulation Study
4.1 Settings and Benchmarks
In this section, we conduct numerical studies to assess the validity, efficiency, and robustness of DEFUSE and compare it with a broad set of existing methods. Our simulation studies include various designs and data generation settings introduced as follows. In every scenario, we consider with fully observed (in ) and possibly missing (in ).
(I) Binary and mixture linear .
In this setting, we first generate binary from the bernoulli distribution with probability , and then generate from the following mixture Gaussian:
where represents the identity matrix. With and , generate
where is an independent noise term. The hyperparameter is set to range from to characterizing the dependence of on the other variables. The sample sizes of the three data sets are set to be , , and with the hyperparameter .
(II) Binary and nonlinear .
We generate in the same way as in (I) but generate through a nonlinear model: and , generate
where is an independent noise. Set , , and .
(III) Continuous and linear model of .
First, we generate independently from the uniform distribution on , and then generate the rest of by
The following linear model is used to simulate the response :
The hyperparameter is set to range from to characterizing the association between and conditional on . The sample sizes of the three data sets are set to be , and with the hyperparameter .
(IV) Linear regression with multiple BM scenario.
First, we generate in the same way as in (III). In this case, set , , and . The sample sizes of the four data sets are set to be and .
In the settings (I) and (II), the outcome model is the logistic model for . In (II), the dependence of on is quadratic, which may make the imputation models subject to model misspecification. We also include (III) and (IV) with linear models for to facilitate the comparison with a broader set of existing methods for BM regression. In (I) and (III), the hyperparameters and are introduced and varied to study how the efficiency gain of our estimators changes according to the dependence between the missing and the observed variables as well as to the sample size ratio between and . In (IV), the hyperparameters and are fixed, and the more complicated multiple BM structure is brought in. In each scenario, we evaluate and compare the estimators through their bias and mean square errors on each coefficient or averaged over all of them.
Methods under comparison.
We summarize all methods under comparison with some implementation details; see also Table 1. -only: The standard estimator obtained using data only. DEFUSE1: Our proposed estimator based on and data. DEFUSE: Our final estimator . MICE: Multiple imputation by chained equations with R package mice (van Buuren and Groothuis-Oudshoorn,, 2011). MBI: The multiple blockwise imputation method proposed by Xue and Qu, (2021) combining and data. HTLGMM: The data fusion approach proposed by Zhao et al., (2023) based on the generalized method of moments. SSB: The SS and BM fusion method proposed by Song et al., (2024) using the , and samples. Benchmarks MBI, HTLGMM, and SSB are all implemented under our low-dimensional regression settings without using any sparse penalties. SemiEff: The classic semiparametric efficient estimator constructed following Robins et al., (1994). SSL: The SS estimator using and (Gronsbell et al.,, 2022, e.g.), with its imputation model for constructed through natural spline regression.
| Method | Nonlinear model | Single BM | Multiple BM | Leverage |
| DEFUSE1 | – | |||
| DEFUSE | ||||
| MICE | – | |||
| MBI | – | – | ||
| SSL | – | – | ||
| HTLGMM | – | – | ||
| SSB | – | |||
| -only | – | – | – | |
| SemiEff | – | – |
For the conditional distribution of used in multiple approaches, we carry out parametric regression to learn its conditional mean function and the standard method of moment to estimate its conditional covariance matrix as described in Remark 1. In (I) and (II), we fit Gaussian linear models for stratified by the binary . In (III) and (IV), we fit Gaussian linear regression for . In settings (I), (III), and (IV), our parametric model for is correctly specified while it is wrong under (II) due to the quadratic effects of . We also fit parametric models for imputation of . For the data-adaptive calibration functions used in DEFUSE, we set and in all settings.
4.2 Comparison with existing methods
In settings (I) and (III), we evaluate the accuracy and efficiency of the included estimators with a fixed in (I) and in (III), and the sample size ratio . The results are summarized in Table 2 for setting (I) and Table 4 for (III). These two tables present the relative efficiency (RE) to the -only estimator defined as the inverse ratio of the average mean squared errors (MSE) over the coefficients . They also include the average mean absolute bias overall coefficients.
All methods except MICE produce small bias and valid estimation. Compared to recent BM data fusion methods, DEFUSE1 attains significantly higher efficiency than HTLGMM in the logistic model setting (I) and higher efficiency than MBI, SSB, and HTLGMM in the linear setting (III). For example, the RE of DEFUSE1 is around larger than that of MBI in setting (III) with . This improvement becomes even larger when grows. As another example, DEFUSE1 is more efficient than HTLGMM in the logistic setting (I) with . As seen from Table 6, our method also attains better performance over both MBI and SSB in setting (IV) with the linear outcome model and multiple BM data structure. For instance, DEFUSE1 has around higher RE than MBI and SSB on in setting (IV).
Meanwhile, DEFUSE1 shows close REs to the classic semiparametric efficient estimator (SemiEff) in both settings (I) and (III) with a correct model for . This finite-sample result is consistent with our Corollary 2 and Remark 7 that when is consistent with the truth, DEFUSE1 will be asymptotically equivalent to SemiEff. Moreover, through incorporating the large sample, DEFUSE achieves around improved average efficiency over DEFUSE1 in all settings. This agrees with our Corollary 3. Such improvements will be demonstrated and discussed more comprehensively and carefully in Section 4.3. We also notice that the standard SSL method has no efficiency gain over the -only estimator in settings (I) and (III) because it only incorporates , which has been shown to contribute zero improvement when the outcome model for is correctly specified (Chakrabortty and Cai,, 2018). This finding also corresponds to Remark 3 that reversing our Steps 2 and 3 in Algorithm 1 tends to result in less efficient estimator.
In Tables 3 and 5, we present the RE (to -only) on each coefficient in the simulation setting (II) with a nonlinear model for not captured by the parametric conditional model. To demonstrate the importance of properly specifying the calibration function , we take and include two choices on for the construction of DEFUSE1: (1) and (2) . Choice only involves an “intercept” term for calibration and is actually relevant to proposals in recent work like Miao et al., (2023). Interestingly, DEFUSE1 with outperforms both SemiEff and DEFUSE1 with . For example, on , DEFUSE1 with has around higher RE than both SemiEff and DEFUSE1 with . This demonstrates that our new data-adaptive calibration approach can effectively improve the estimation efficiency over the classic SemiEff when the imputation model is wrongly specified or of poor quality. To realize this advantage, it is important to specify the calibration function in a proper way, which can be seen by comparing DEFUSE1 under the two different choices on in this setting.
| Methods | ratio = 2 | ratio = 3 | ratio = 4 | ratio = 5 | ||||
| RE | Bias | RE | Bias | RE | Bias | RE | Bias | |
| DEFUSE1 | 1.74 | 0.011 | 1.90 | 0.012 | 2.15 | 0.015 | 2.23 | 0.015 |
| DEFUSE | 1.88 | 0.007 | 2.09 | 0.010 | 2.45 | 0.012 | 2.56 | 0.012 |
| MICE | 0.93 | 0.190 | 1.00 | 0.214 | 1.18 | 0.229 | 1.30 | 0.240 |
| SSL | 1.01 | 0.020 | 1.01 | 0.020 | 1.01 | 0.020 | 1.01 | 0.020 |
| HTLGMM | 1.42 | 0.029 | 1.46 | 0.033 | 1.52 | 0.035 | 1.54 | 0.036 |
| -only | 1 | 0.019 | 1 | 0.019 | 1 | 0.019 | 1 | 0.019 |
| SemiEff | 1.76 | 0.010 | 1.92 | 0.010 | 2.18 | 0.012 | 2.26 | 0.012 |
| Methods | ratio = 2 | ratio = 3 | ratio = 4 | ratio = 5 | ||||
| RE | Bias | RE | Bias | RE | Bias | RE | Bias | |
| SemiEff | 1.88 | 0.020 | 2.22 | 0.010 | 2.52 | 0.016 | 2.63 | 0.019 |
| DEFUSE1 | 1.91 | 0.018 | 2.26 | 0.018 | 2.57 | 0.024 | 2.70 | 0.022 |
| DEFUSE1 | 2.09 | 0.013 | 2.51 | 0.021 | 2.98 | 0.022 | 3.23 | 0.027 |
| MICE | 2.22 | 0.103 | 2.72 | 0.135 | 3.66 | 0.151 | 4.11 | 0.167 |
| SSL | 1.04 | 0.47 | 1.06 | 0.47 | 1.04 | 0.45 | 1.05 | 0.046 |
| HTLGMM | 1.44 | 0.038 | 1.63 | 0.037 | 1.78 | 0.042 | 1.85 | 0.046 |
| Methods | ratio = 2 | ratio = 3 | ratio = 4 | ratio = 5 | ||||
| RE | Bias | RE | Bias | RE | Bias | RE | Bias | |
| DEFUSE1 | 1.32 | 0.006 | 1.38 | 0.007 | 1.39 | 0.007 | 1.42 | 0.007 |
| DEFUSE | 1.41 | 0.004 | 1.51 | 0.004 | 1.56 | 0.004 | 1.62 | 0.004 |
| MICE | 0.06 | 0.294 | 0.04 | 0.381 | 0.03 | 0.453 | 0.02 | 0.506 |
| MBI | 1.11 | 0.006 | 1.15 | 0.007 | 1.14 | 0.007 | 1.16 | 0.007 |
| SSL | 0.98 | 0.005 | 0.98 | 0.005 | 0.98 | 0.005 | 0.98 | 0.005 |
| HTLGMM | 1.15 | 0.005 | 1.21 | 0.005 | 1.20 | 0.006 | 1.22 | 0.006 |
| SSB | 0.95 | 0.007 | 1.03 | 0.009 | 1.07 | 0.006 | 1.11 | 0.006 |
| -only | 1 | 0.005 | 1 | 0.005 | 1 | 0.005 | 1 | 0.005 |
| SemiEff | 1.32 | 0.006 | 1.38 | 0.007 | 1.39 | 0.007 | 1.42 | 0.007 |
| Method | |||||
|---|---|---|---|---|---|
| SemiEff | 4.48 | 3.84 | 2.86 | 0.99 | 1.00 |
| DEFUSE1 | 4.44 | 3.99 | 3.12 | 0.98 | 0.96 |
| DEFUSE1 | 4.87 | 5.11 | 3.90 | 1.23 | 1.06 |
| MICE | 4.86 | 10.01 | 4.92 | 0.18 | 0.58 |
| SSL | 1.01 | 1.08 | 1.00 | 0.93 | 1.25 |
| HTLGMM | 2.41 | 2.80 | 1.90 | 1.06 | 1.09 |
| Method | |||||
|---|---|---|---|---|---|
| DEFUSE1 | 1.60 | 1.78 | 2.39 | 1.49 | 1.67 |
| DEFUSE | 1.69 | 1.83 | 2.55 | 1.51 | 1.68 |
| MICE | 0.52 | 0.63 | 0.69 | 0.20 | 0.22 |
| MBI | 1.06 | 1.09 | 1.06 | 0.95 | 1.01 |
| SSB | 1.07 | 1.12 | 1.51 | 0.60 | 0.81 |
| SSL | 1.01 | 1.00 | 1.00 | 0.99 | 1.02 |
4.3 Patterns of relative efficiency
In this section, we present more detailed results in settings (I) and (III) to assess DEFUSE under different values of and quantifying the dependence of on . We only present results on the coefficients . The results on other coefficients are similar and can be found in Appendix. We focus on the RE of DEFUSE1 to the -only estimator as well as the RE of DEFUSE to DEFUSE1.
In Figures 3 and 5, we present the REs of DEFUSE1 to -only. A larger efficiency gain of DEFUSE1 over -only can be observed with an increasing sample size ratio ; see also Tables 2 and 4. This finding agrees with our Corollary 1 that monotonically increases with . On corresponding to the partially missing , the efficiency gain of DEFUSE1 tends to be higher with an increasing dependence parameter . Intuitively, when , will be independent from and the sample with only observed cannot provide any information on . Consequently, incorporating could not help much on whose score function is largely dependent on . On the other hand, under a large , observed on can be informative to the score function of and improve the estimation efficiency on it. Interestingly, for and corresponding to covariates in observed on , increasing tends to cause a lower efficiency gain of our method, which is opposite to the pattern of REs on . For example, in setting (III) with , the RE of DEFUSE1 on decrease from to when changes from to ; see Figure 5. This is because when becomes larger, the score functions of tend to be more dependent on , which makes the missingness of on more impactful.
In Figures 4 and 6, we explore the RE trends of DEFUSE to DEFUSE1 under different sample size ratios and weights . Similarly, we find that a larger results in more efficiency gain of DEFUSE over DEFUSE1, which agrees with Corollary 3 that will increase with when the nuisance and are of good quality. Also, on corresponding to the BM covaraite , the RE of DEFUSE to DEFUSE1 tends to become larger when the dependence of on characterized by gets larger. Nevertheless, the overall improvements of DEFUSE over DEFUSE1 is relatively small on . Interestingly, on and , we find that the RE first increases and then decreases with being larger. Future theoretical analysis is needed to help understanding and explaining this non-monotonic trend better.




5 Heart Disease Risk Modeling with MIMIC-III
5.1 Extension on Auxiliary Covariates
We begin with introducing a natural extension of our framework to include auxiliary features, which will be used in our real-world study. Consider the scenario with covariates consisting of the primary risk factors and auxiliary or surrogate features . Instead of the whole , the focus is still on defined by the population equation (2). The missing structure on remains the same as described in Section 1.2, i.e., one or multiple subsets of covariates partially missing on the labeled sample and being unlabeled on with complete . Meanwhile, suppose to be complete on all subjects.
Such auxiliary features are commonly used in practice. For instance, in EMR linked biobank study, the interest lies in the relationship between certain genetic variants and disease status . is taken as EMR proxies (e.g., relevant diagnostic codes) for and acts as auxiliary features not included in the genetic risk model. In clinical or epidemiological studies, is the treatment or key risk factors, is a long-term outcome, and can be some early endpoints or surrogate outcomes (e.g., tumor response rates) occurring and observed post the baseline with only . In this case, one is interested in to make clinical decision at the baseline without . In both examples, the auxiliary could still be informative to and and, thus, included to boost the statistical efficiency.
Our framework can be naturally extended to accommodate this setting. For the construction described in Section 2 and summarized in Algorithm 1, the only change to make is to allow the inclusion of in the nuisance imputation models and , as well as in the calibration functions and . In this case, could often be of high-dimensionality as considered in Zhou et al., (2024) as well as our real example. The theoretical results established in Section 3 can also be applied to this setting. Despite the association of with both and , including in the nuisance imputation models will not create bias thanks to our constructions like (7) enabling automatic bias correction with respect to the nuisance estimators. It is also important to note that our data-adaptive control function approach can be particularly useful in the presence of erroneous nuisance estimators with high-dimensional , which will be illustrated though the real example in this section.
5.2 Setup
To illustrate the utility of DEFUSE, we apply it to MIMIC-III (Medical Information Mart for Intensive Care III; Johnson et al., (2016)), a large database comprising de-identified electronic health records (EMRs) of over forty thousand patients associated with their stay in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012. The database includes various types of information such as demographics, vital sign measurements made at the bedside, laboratory test results, procedures, medications, caregiver notes, imaging reports, and mortality in and out of the hospital. As EMRs include large and diverse patient populations and cover comprehensive phenotypes, they have been widely used by researchers to identify underlying risk factors for certain disease conditions. Our goal is to make statistical inferences on the risk prediction model for heart diseases (HD) against some potential risk factors related to the health condition of a subject.
Our response of interest is the HD status identified from the manual chart reviewing labels obtained in a previous study on MIMIC-III data (Gehrmann et al.,, 2018); see their paper for the detailed definition of the phenotype HD. We greatly thank the authors for making their gold-standard labels accessible. For , we include age, encounter of the International Classification of Diseases (ICD) codes of low high-density lipoproteins (HDL), ICD encounter of type II diabetes, and several HD-related laboratory (lab) variables selected using KESER (Hong et al.,, 2021), an online tool for extracting clinical concepts and features relevant to a given phenotype. For the convenience of downstream risk modeling, we derive a lab risk score (LRS) through weighting and combining the selected lab variables with the main ICD code of HD as a surrogate outcome on the large sample. Since the LRS is derived with a large sample, it has a negligible estimation uncertainty and can be viewed as given. See the list of selected lab variables and details about the LRS in Appendix.
Noticing that each lab variable is not observed on a particular fraction of patients, we separately consider two settings of missingness: (i) a single BM task with one BM covariate taken as the derived LRS; (ii) a multiple BM task with two BM lab variables thyroid-stimulating hormone (TSH) and hypochromia (HPO), with joint missingness on , TSH-only missingness on , HPO-only missing on . On task (i), there are labeled subject with complete observation of , labeled subjects with BM covariates, and unlabeled subjects with complete . Similar to the simulation, we compare our approaches to four applicable benchmarks to this case, including -only, SSL, Semieff, and HTLGMM. We do not include MICE since it tends to result in excessive bias, as seen from the simulation results. For (ii), there are labeled subject with complete observation of , , , and for three types of missing, and .
To assist data-fusion, we include auxiliary features as introduced in Section 5.1. With all codified features including the ICD and procedure codes in EMR, we apply a simple pre-screening procedure with the ICD count of HD as a surrogate outcome against around all the EMR features on the whole sample, to select features and include them in for the downstream risk model estimation. For the implementation of DEFUSE, we follow the same strategy as our simulation with the conditional models constructed separately through kernel machine (KM) and Lasso with their hyper-parameters tuned by cross-validation. Similar to our simulation design, the calibration functions are set as and and we also include DEFUSE1 with for comparison with existing calibration strategies (Miao et al.,, 2023, e.g.). To ensure fair comparison, we use the same machine learning estimators for missing variable imputation in all the benchmark methods if needed.
5.3 Results
Table 7 reports the relative efficiency (RE) of the data-fusing estimators to -only in the single BM task (i). DEFUSE achieves the highest REs on all coefficients. For example, in estimating the HDL coefficient, our method reduces more than variance compared to -only and its RE to HTLGMM is around . Interestingly, under both the KM and Lasso implementation, DEFUSE1 with the calibration basis attains smaller estimation variance on the partially missing predictor LRS compared to SemiEff and DEFUSE1 with while they all have close performance on the other predictors. In specific, with Lasso imputation, DEFUSE1 with has improved efficiency over -only while either SemiEff or DEFUSE1 with has no improvement over -only. Similarly, when using KM, DEFUSE1 with attains a higher RE than SemiEff and DEFUSE1 with . This demonstrates not only the importance of data-adaptive calibration on the erroneous machine learning estimators, but also the effectiveness of our newly proposed control function strategy in Section 2.2 that is more general and sophisticated than the simple one with . In addition, on LRS, DEFUSE with the SSL step shows a further improvement over DEFUSE1 with 6-7% higher efficiency while such an improvement is not significant on the other coefficients.
Table 8 provides a summary of the estimated coefficients as well as the bootstrap standard errors and -values on the single BM task, with the nuisance imputation models fitted using KM. Through the output of DEFUSE, one can conclude that given other risk factors, a higher risk of HD is associated with low HDL at the level as well as type II diabetes in the level . Notably, with -only and SSL, both the effects of HDL and diabetes are shown to be insignificant even in the level and diabetes is not significant in HTLGMM. This suggests that DEFUSE is more powerful than these methods to discover both signals. Interestingly, it is well-documented in biomedical literature on the associations between heart disease and diabetes (Haffner,, 2000; Peters et al.,, 2014, e.g.) as well as low HDL (Després et al.,, 2000; Rader and Hovingh,, 2014, e.g.), which supports the findings of DEFUSE.
At last, Table 9 summarizes the RE to the -only estimator in the multiple BM scenario introduced above. DEFUSE attains more than efficiency improvement over -only on all coefficients and more than variance reduction on that of age, which is again more significant than SSL. One can find the summary table of the estimates and -values in this scenario in Appendix.
| Method | Age | HDL | Diabetes | LRS |
| SemiEff (KM) | 2.15 | 2.21 | 2.20 | 1.08 |
| DEFUSE1 (KM) | 2.15 | 2.21 | 2.20 | 1.08 |
| DEFUSE1 (KM) | 2.16 | 2.22 | 2.20 | 1.17 |
| DEFUSE (KM) | 2.19 | 2.24 | 2.22 | 1.24 |
| SemiEff (Lasso) | 2.14 | 2.20 | 2.19 | 0.97 |
| DEFUSE1 (Lasso) | 2.14 | 2.20 | 2.19 | 1.00 |
| DEFUSE1 (Lasso) | 2.14 | 2.20 | 2.19 | 1.10 |
| DEFUSE (Lasso) | 2.17 | 2.23 | 2.22 | 1.16 |
| SSL | 1.07 | 0.99 | 1.00 | 1.00 |
| HTLGMM | 1.76 | 1.55 | 1.56 | 1.02 |
| Method | Age | HDL | Diabetes | LRS | ||||
|---|---|---|---|---|---|---|---|---|
| -value | -value | -value | -value | |||||
| -only | 0.002 | 0.136 | 0.509 | 0.032 | ||||
| SemiEff | 0.001 | 0.093 | 0.047 | 0.004 | ||||
| DEFUSE1 | 0.001 | 0.088 | 0.051 | 0.002 | ||||
| DEFUSE | 0.001 | 0.07 | 0.05 | 0.001 | ||||
| SSL | 0.001 | 0.140 | 0.480 | 0.032 | ||||
| HTLGMM | 0.001 | 0.046 | 0.174 | 0.029 | ||||
| Method | Age | HDL | Diabetes | TSH | Hypochromia |
| DEFUSE1 | 2.41 | 1.29 | 1.30 | 1.71 | 1.27 |
| DEFUSE | 2.42 | 1.29 | 1.31 | 1.82 | 1.28 |
| SSL | 1.00 | 0.99 | 1.06 | 0.94 | 1.13 |
6 Discussion
In this paper, we develop DEFUSE, a novel approach for robust and efficient data-fusion in the presence of blockwise missing covariates and large unlabeled samples. Its validity and relative efficiency compared to a comprehensive set of existing methods are justified through theoretical, numerical, and real-world studies. The principle of our construction strategy is to leverage the general and powerful theory on semiparametric efficient estimation while incorporating the key data-adaptive calibration and optimal linear allocation procedures to maintain robustness and effectiveness to a more realistic scenario that the nuisance distributional models are imperfect or problematic. We shall point out the limitation and future direction of our work.
Our linear allocation strategy for the multiple BM scenario does not achieve semiparametric efficiency. We notice that the general canonical gradient method proposed by Li and Luedtke, (2023) could be leveraged to further improve in this direction, even with non-nested sets of observed covariates in . Nevertheless, it is still an open question about how to incorporate our data-adaptive calibration approach and the SS setting into this more complicated framework. In the simulation study, we adopt the parametric and kernel methods to construct the conditional models and . As was pointed out, DEFUSE allows the use of more complex tools like generative adversarial models to estimate the unknown conditional distributions, which could achieve better performance for relatively high-dimensional missing covariates. However, care must be taken to avoid over-fitting and overly high computational costs.
Also, our current method requires sample that may not be accessible in certain scenarios (Song et al.,, 2024, e.g.). It is not hard to see that (2) can actually be identified and constructed with the and samples as long as the union set of the observed covariates in ’s is complete. It is interesting to generalize our method to accommodate this setting without observation of . One possible way is to first utilize a part of data sets to derive a preliminary estimator that is asymptotically normal but not efficient, and then implement similar steps to Algorithm 1 to reduce its variance. Other more complicated missing structures, e.g., single or multiple missing blocks in the unlabeled sample may also warrant future research.
We notice a comprehensive set of recent literature in addressing covariate shift of (Liu et al.,, 2023; Qiu et al.,, 2023, e.g.), label or model shift of (Li et al., 2023a, ; Zhao et al.,, 2023, e.g.), as well as high-dimensionality (Xue et al.,, 2021; Song et al.,, 2024, e.g.) in data fusion under various types of missingness. DEFUSE can also be generalized to address these practical issues. For high-dimensional sparse regression model of , we can use debiased Lasso (Zhang and Zhang,, 2014; Van de Geer et al.,, 2014; Javanmard and Montanari,, 2014, e.g.) in Step 1 of Algorithm 1 to derive the preliminary estimator that is -consistent and asymptotically normal. Then the remaining Steps 2 and 3 can be implemented in a similar way as Algorithm 1. Since our main theorems only require the convergence of the nuisance models, construction procedures of our nuisance parameters could naturally incorporate sparse regularization to accommodate high-dimensionality without introducing excessive bias. For potential covariate shift across the , and samples, one can introduce density ratio (propensity score) models for adjustment (Qiu et al.,, 2023; Liu et al.,, 2023, e.g.) based on our current MCAR construction. This can result in doubly robust forms requiring stronger assumption on the correctness and convergence of the nuisance models. For the conditional model shift of , adaptive data fusion under our setting can be challenging, especially for statistical inference. We notice some related recent literature like Han et al., (2021) and Li et al., 2023a , and leave this open problem to future research.
References
- Angelopoulos et al., (2023) Angelopoulos, A. N., Duchi, J. C., and Zrnic, T. (2023). Ppi++: Efficient prediction-powered inference. arXiv preprint arXiv:2311.01453.
- Athey et al., (2019) Athey, S., Tibshirani, J., and Wager, S. (2019). Generalized random forests. The Annals of Statistics, 47(2):1148–1178.
- Azriel et al., (2022) Azriel, D., Brown, L. D., Sklar, M., Berk, R., Buja, A., and Zhao, L. (2022). Semi-supervised linear regression. Journal of the American Statistical Association, 117(540):2238–2251.
- Bycroft et al., (2018) Bycroft, C., Freeman, C., Petkova, D., Band, G., Elliott, L. T., Sharp, K., Motyer, A., Vukcevic, D., Delaneau, O., O’Connell, J., et al. (2018). The uk biobank resource with deep phenotyping and genomic data. Nature, 562(7726):203–209.
- Caflisch, (1998) Caflisch, R. E. (1998). Monte carlo and quasi-monte carlo methods. Acta numerica, 7:1–49.
- Cai and Guo, (2020) Cai, T. T. and Guo, Z. (2020). Semisupervised inference for explained variance in high dimensional linear regression and its applications. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 82(2):391–419.
- Castro et al., (2022) Castro, V. M., Gainer, V., Wattanasin, N., Benoit, B., Cagan, A., Ghosh, B., Goryachev, S., Metta, R., Park, H., Wang, D., et al. (2022). The mass general brigham biobank portal: an i2b2-based data repository linking disparate and high-dimensional patient data to support multimodal analytics. Journal of the American Medical Informatics Association, 29(4):643–651.
- Chakrabortty and Cai, (2018) Chakrabortty, A. and Cai, T. (2018). Efficient and adaptive linear regression in semi-supervised settings. The Annals of Statistics, 46(4):1541–1572.
- Chen et al., (2008) Chen, X., Hong, H., and Tarozzi, A. (2008). Semiparametric efficiency in gmm models with auxiliary data.
- Chernozhukov et al., (2018) Chernozhukov, V., Chetverikov, D., Demirer, M., Duflo, E., Hansen, C., Newey, W., and Robins, J. (2018). Double/debiased machine learning for treatment and structural parameters. The Econometrics Journal, 21(1):C1–C68.
- Chernozhukov et al., (2016) Chernozhukov, V., Chetverikov, D., Demirer, M., Duflo, E., Hansen, C., and Newey, W. K. (2016). Double machine learning for treatment and causal parameters. Technical report, cemmap working paper.
- Deng et al., (2023) Deng, S., Ning, Y., Zhao, J., and Zhang, H. (2023). Optimal and safe estimation for high-dimensional semi-supervised learning. Journal of the American Statistical Association, pages 1–12.
- Després et al., (2000) Després, J.-P., Lemieux, I., Dagenais, G.-R., Cantin, B., and Lamarche, B. (2000). Hdl-cholesterol as a marker of coronary heart disease risk: the quebec cardiovascular study. Atherosclerosis, 153(2):263–272.
- Gan and Liang, (2023) Gan, F. and Liang, W. (2023). Prediction de-correlated inference. arXiv preprint arXiv:2312.06478.
- Gehrmann et al., (2018) Gehrmann, S., Dernoncourt, F., Li, Y., Carlson, E. T., Wu, J. T., Welt, J., Foote Jr, J., Moseley, E. T., Grant, D. W., Tyler, P. D., et al. (2018). Comparing deep learning and concept extraction based methods for patient phenotyping from clinical narratives. PloS one, 13(2):e0192360.
- Gronsbell et al., (2022) Gronsbell, J., Liu, M., Tian, L., and Cai, T. (2022). Efficient evaluation of prediction rules in semi-supervised settings under stratified sampling. Journal of the Royal Statistical Society: Series B: Statistical Methodology.
- Haffner, (2000) Haffner, S. M. (2000). Coronary heart disease in patients with diabetes.
- Han et al., (2021) Han, L., Hou, J., Cho, K., Duan, R., and Cai, T. (2021). Federated adaptive causal estimation (face) of target treatment effects. arXiv preprint arXiv:2112.09313.
- Hatt et al., (2022) Hatt, T., Berrevoets, J., Curth, A., Feuerriegel, S., and van der Schaar, M. (2022). Combining observational and randomized data for estimating heterogeneous treatment effects. arXiv preprint arXiv:2202.12891.
- Hong et al., (2021) Hong, C., Rush, E., Liu, M., Zhou, D., Sun, J., Sonabend, A., Castro, V. M., Schubert, P., Panickan, V. A., Cai, T., et al. (2021). Clinical knowledge extraction via sparse embedding regression (keser) with multi-center large scale electronic health record data. NPJ digital medicine, 4(1):1–11.
- Javanmard and Montanari, (2014) Javanmard, A. and Montanari, A. (2014). Confidence intervals and hypothesis testing for high-dimensional regression. The Journal of Machine Learning Research, 15(1):2869–2909.
- Jin and Rothenhäusler, (2023) Jin, Y. and Rothenhäusler, D. (2023). Modular regression: Improving linear models by incorporating auxiliary data. Journal of Machine Learning Research, 24(351):1–52.
- Johns, (1988) Johns, M. V. (1988). Importance sampling for bootstrap confidence intervals. Journal of the American Statistical Association, 83(403):709–714.
- Johnson et al., (2016) Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L.-w. H., Feng, M., Ghassemi, M., Moody, B., Szolovits, P., Anthony Celi, L., and Mark, R. G. (2016). Mimic-iii, a freely accessible critical care database. Scientific data, 3(1):1–9.
- Kawakita and Kanamori, (2013) Kawakita, M. and Kanamori, T. (2013). Semi-supervised learning with density-ratio estimation. Machine learning, 91(2):189–209.
- Kovvali, (2022) Kovvali, N. (2022). Theory and applications of Gaussian quadrature methods. Springer Nature.
- Kundu and Chatterjee, (2023) Kundu, P. and Chatterjee, N. (2023). Logistic regression analysis of two-phase studies using generalized method of moments. Biometrics, 79(1):241–252.
- (28) Li, S., Gilbert, P. B., and Luedtke, A. (2023a). Data fusion using weakly aligned sources. arXiv preprint arXiv:2308.14836.
- Li and Luedtke, (2023) Li, S. and Luedtke, A. (2023). Efficient estimation under data fusion. Biometrika, 110(4):1041–1054.
- (30) Li, Y., Yang, H., Yu, H., Huang, H., and Shen, Y. (2023b). Penalized estimating equations for generalized linear models with multiple imputation. The Annals of Applied Statistics, 17(3):2345–2363.
- Liu et al., (2023) Liu, M., Zhang, Y., Liao, K. P., and Cai, T. (2023). Augmented transfer regression learning with semi-non-parametric nuisance models. Journal of Machine Learning Research, 24(293):1–50.
- The All of Us Research Program Investigators, (2019) The All of Us Research Program Investigators (2019). The “all of us” research program. New England Journal of Medicine, 381(7):668–676.
- Miao et al., (2023) Miao, J., Miao, X., Wu, Y., Zhao, J., and Lu, Q. (2023). Assumption-lean and data-adaptive post-prediction inference. arXiv preprint arXiv:2311.14220.
- Mirza and Osindero, (2014) Mirza, M. and Osindero, S. (2014). Conditional generative adversarial nets. arXiv preprint arXiv:1411.1784.
- Negahban et al., (2012) Negahban, S. N., Ravikumar, P., Wainwright, M. J., Yu, B., et al. (2012). A unified framework for high-dimensional analysis of -estimators with decomposable regularizers. Statistical Science, 27(4):538–557.
- Peters et al., (2014) Peters, S. A., Huxley, R. R., and Woodward, M. (2014). Diabetes as risk factor for incident coronary heart disease in women compared with men: a systematic review and meta-analysis of 64 cohorts including 858,507 individuals and 28,203 coronary events. Diabetologia, 57:1542–1551.
- Qiu et al., (2023) Qiu, H., Tchetgen, E. T., and Dobriban, E. (2023). Efficient and multiply robust risk estimation under general forms of dataset shift. arXiv preprint arXiv:2306.16406.
- Rader and Hovingh, (2014) Rader, D. J. and Hovingh, G. K. (2014). Hdl and cardiovascular disease. The Lancet, 384(9943):618–625.
- Robins et al., (1994) Robins, J. M., Rotnitzky, A., and Zhao, L. P. (1994). Estimation of regression coefficients when some regressors are not always observed. Journal of the American statistical Association, 89(427):846–866.
- Schmutz et al., (2022) Schmutz, H., Humbert, O., and Mattei, P.-A. (2022). Don’t fear the unlabelled: safe semi-supervised learning via debiasing. In The Eleventh International Conference on Learning Representations.
- Shi et al., (2023) Shi, X., Pan, Z., and Miao, W. (2023). Data integration in causal inference. Wiley Interdisciplinary Reviews: Computational Statistics, 15(1):e1581.
- Song et al., (2023) Song, S., Lin, Y., and Zhou, Y. (2023). A general m-estimation theory in semi-supervised framework. Journal of the American Statistical Association, pages 1–11.
- Song et al., (2024) Song, S., Lin, Y., and Zhou, Y. (2024). Semi-supervised inference for block-wise missing data without imputation. Journal of Machine Learning Research, 25(99):1–36.
- Sudlow et al., (2015) Sudlow, C., Gallacher, J., Allen, N., Beral, V., Burton, P., Danesh, J., Downey, P., Elliott, P., Green, J., Landray, M., et al. (2015). Uk biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS medicine, 12(3):e1001779.
- Tian et al., (2007) Tian, L., Cai, T., Goetghebeur, E., and Wei, L. (2007). Model evaluation based on the sampling distribution of estimated absolute prediction error. Biometrika, 94(2):297–311.
- van Buuren and Groothuis-Oudshoorn, (2011) van Buuren, S. and Groothuis-Oudshoorn, K. (2011). mice: Multivariate imputation by chained equations in r. Journal of Statistical Software, 45(3):1–67.
- Van de Geer et al., (2014) Van de Geer, S., Bühlmann, P., Ritov, Y., Dezeure, R., et al. (2014). On asymptotically optimal confidence regions and tests for high-dimensional models. The Annals of Statistics, 42(3):1166–1202.
- Van der Vaart, (2000) Van der Vaart, A. W. (2000). Asymptotic statistics, volume 3. Cambridge university press.
- Verma et al., (2023) Verma, A., Huffman, J. E., Rodriguez, A., Conery, M., Liu, M., Ho, Y.-L., Kim, Y., Heise, D. A., Guare, L., Panickan, V. A., et al. (2023). Diversity and scale: genetic architecture of 2,068 traits in the va million veteran program. medRxiv.
- Wang et al., (2024) Wang, X., Liu, M., Nogues, I.-E., Chen, T., Xiong, X., Bonzel, C.-L., Zhang, H., Hong, C., Xia, Y., Dahal, K., et al. (2024). Heterogeneous associations between interleukin-6 receptor variants and phenotypes across ancestries and implications for therapy. Scientific Reports, 14(1):8021.
- White et al., (2011) White, I. R., Royston, P., and Wood, A. M. (2011). Multiple imputation using chained equations: issues and guidance for practice. Statistics in medicine, 30(4):377–399.
- Wolfson et al., (2010) Wolfson, M., Wallace, S. E., Masca, N., Rowe, G., Sheehan, N. A., Ferretti, V., LaFlamme, P., Tobin, M. D., Macleod, J., Little, J., et al. (2010). DataSHIELD: resolving a conflict in contemporary bioscience performing a pooled analysis of individual-level data without sharing the data. International journal of epidemiology, 39(5):1372–1382.
- Wu and Liu, (2023) Wu, D. and Liu, M. (2023). Robust and efficient semi-supervised learning for ising model. arXiv preprint arXiv:2311.15031.
- Xue et al., (2021) Xue, F., Ma, R., and Li, H. (2021). Statistical inference for high-dimensional linear regression with blockwise missing data. arXiv preprint arXiv:2106.03344.
- Xue and Qu, (2021) Xue, F. and Qu, A. (2021). Integrating multisource block-wise missing data in model selection. Journal of the American Statistical Association, 116(536):1914–1927.
- Yang and Ding, (2019) Yang, S. and Ding, P. (2019). Combining multiple observational data sources to estimate causal effects. Journal of the American Statistical Association.
- Yu et al., (2020) Yu, G., Li, Q., Shen, D., and Liu, Y. (2020). Optimal sparse linear prediction for block-missing multi-modality data without imputation. Journal of the American Statistical Association, 115(531):1406–1419.
- Zengini et al., (2018) Zengini, E., Hatzikotoulas, K., Tachmazidou, I., Steinberg, J., Hartwig, F. P., Southam, L., Hackinger, S., Boer, C. G., Styrkarsdottir, U., Gilly, A., et al. (2018). Genome-wide analyses using uk biobank data provide insights into the genetic architecture of osteoarthritis. Nature genetics, 50(4):549–558.
- Zhang and Zhang, (2014) Zhang, C.-H. and Zhang, S. S. (2014). Confidence intervals for low dimensional parameters in high dimensional linear models. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 76(1):217–242.
- Zhang et al., (2023) Zhang, H., Li, Y., Lu, W., and Lin, Q. (2023). On the optimality of misspecified kernel ridge regression. In International Conference on Machine Learning, pages 41331–41353. PMLR.
- Zhang and Bradic, (2022) Zhang, Y. and Bradic, J. (2022). High-dimensional semi-supervised learning: in search of optimal inference of the mean. Biometrika, 109(2):387–403.
- Zhao et al., (2023) Zhao, R., Kundu, P., Saha, A., and Chatterjee, N. (2023). Heterogeneous transfer learning for building high-dimensional generalized linear models with disparate datasets. arXiv preprint arXiv:2312.12786.
- Zhou et al., (2024) Zhou, D., Liu, M., Li, M., and Cai, T. (2024). Doubly robust augmented model accuracy transfer inference with high dimensional features. Journal of the American Statistical Association, (just-accepted):1–26.