A MINI-BATCH METHOD For SOLVING NONLINEAR PDES WITH GAUSSIAN PROCESSES
Abstract.
Gaussian processes (GPs) based methods for solving partial differential equations (PDEs) demonstrate great promise by bridging the gap between the theoretical rigor of traditional numerical algorithms and the flexible design of machine learning solvers. The main bottleneck of GP methods lies in the inversion of a covariance matrix, whose cost grows cubically concerning the size of samples. Drawing inspiration from neural networks, we propose a mini-batch algorithm combined with GPs to solve nonlinear PDEs. A naive deployment of a stochastic gradient descent method for solving PDEs with GPs is challenging, as the objective function in the requisite minimization problem cannot be depicted as the expectation of a finite-dimensional random function. To address this issue, we employ a mini-batch method to the corresponding infinite-dimensional minimization problem over function spaces. The algorithm takes a mini-batch of samples at each step to update the GP model. Thus, the computational cost is allotted to each iteration. Using stability analysis and convexity arguments, we show that the mini-batch method steadily reduces a natural measure of errors towards zero at the rate of , where is the number of iterations and is the batch size.
1. Introduction
This work develops a mini-batch method based on stochastic proximal algorithms [12, 1, 2] to solve nonlinear partial differential equations (PDEs) with Gaussian processes (GPs). PDEs have been widely used in science, economics, and biology [35, 44]. Since PDEs generally lack explicit solutions, numerical approximations are essential in applications. Standard numerical methods, for instance, finite difference [44] and finite element methods [19], for solving nonlinear PDEs are prone to the curse of dimensions. Recently, to keep pace with scaling problem sizes, machine learning methods, including neural networks [39, 27, 20] and probabilistic algorithms [38, 21, 9], arise a lot of attention. Especially, GPs present a promising approach that integrates the rigorous principles of traditional numerical algorithms with the adaptable framework of machine learning solvers [38, 9, 46, 3]. Our methods are based on the work of [9], which proposes a GP framework for solving general nonlinear PDEs. A numerical approximation of the solution to a nonlinear PDE is viewed as the maximum a posteriori (MAP) estimator of a GP conditioned on solving the PDE at a finite set of sample points. Equivalently, [9] proposes to approximate the unique strong solution of a nonlinear PDE in a reproducing kernel Hilbert space (RKHS) by a minimizer of the following optimal recovery problem
| (1.1) |
where is a small regularization parameter, is the number of samples in the domain, encodes the PDE equation satisfied at the sample, and represents the linear operators in the equation. Let , let be the kernel associated with , and let be the domain of the PDE. Denote by the action of on for fixed and by the action of on . By the representer theorem (see [34, Sec. 17.8]), a solution of (1.1) satisfies , where solves
| (1.2) |
Then, [9] proposes to solve (1.2) by the Gauss–Newton method. We refer readers to [9, 29] for illustrative examples on how to transform a PDE solving task into the minimization problem described in (1.1). The computational cost of the above GP method grows cubically with respect to the number of samples due to the inversion of the covariance matrix . To improve the performance, based on [9], [31] proposes to approximate the solution of a PDE in a function space generated by random Fourier features [37, 49], where the corresponding covariance matrix is a low-rank approximation to . In [29], another low-rank approximation method using sparse GPs is presented, employing a small number of inducing points to summarize observation information. Additionally, [10] introduces an algorithm that efficiently computes the sparse Cholesky factorization of the inverted Gram matrix. Despite their computational cost-saving accomplishments, low-rank approximation methods lack the scalability of neural network-based algorithms. The primary cause for this difference lies in the scaling of parameters in the GP/Kernel method with respect to the number of collocation points, which in turn escalates with the increase in dimensions. Conversely, in neural network models, the parameters are designed to be independent of the number of data points.
Compared to methods based on neural networks [39, 28, 20] , Gaussian Process (GP)/Kernel-based approaches generally offer solid theoretical foundations. Additionally, from a probabilistic viewpoint, GP regressions provide uncertainty quantification. This quantification allows us to establish the confidence intervals of the results and aids in deciding the placement of collocation points. The exploration of how to effectively utilize uncertainty quantification will be a subject for future research. Nevertheless, the GP method typically incurs a computational cost of , where represents the number of collocation points. On the other hand, besides their capacity to model a wide range of functions, neural network methods also excel in scalability, largely due to the use of stochastic gradient descent (SGD) algorithms with mini-batches. This scalability is facilitated by the independence of their data points from the parameters. However, due to the global correlation of the unknowns in (1.2), the direct application of SGD is impeded by the need to solve the inversion of the covariance matrix in equation (1.2). Consequently, devising an effective SGD approach is vital for the GP methodology to reach a level of scalability on par with neural networks. This paper attempts to handle this issue by proposing a mini-batch method inspired by stochastic proximal methods [12, 1, 2]. Rather than pursuing low-rank approximations, we stochastically solve equation (1.1). Each iteration selects a small portion of samples to update the GP model. Hence, we only need to consider the inversion of the covariance matrix of samples in a mini-batch each time. More precisely, we introduce a slack variable and reformulate (1.1) as
| (1.3) |
where is a convex compact set, , and . We note that as , the solutions of (1.3) converge to those of (1.1). The latent variable encodes observations of . Next, we view (1.3) as a stochastic optimization problem
| (1.4) |
where is a uniform random sample taken from the set of the whole samples. Then, we iteratively solve (1.4) by computing the proximal map
| (1.5) |
where contains a mini-batch of samples, is the cardinality of , and is the step size at the step. Denote by the vector containning elements of a vector indexed by . Then, to handle the inifnite dimensional minimization problem in (1), we derive a representer formula for (1) in Section 2 such that
| (1.6) |
where is the identity matrix, , and solves
| (1.7) |
Thus, the computational cost is reduced to in each iteration if we use the standard Cholesky decomposition to compute the inverse of the covariance matrix for solving (1.6) and (1.7). Under assumptions of boundedness of and the weakly convexity of the functions , our convergence analysis shows that the error diminishes at the rate of , where is the number of iterations and is the batch size. (For clarification, it’s important to note that the computational complexity of addressing the inner minimization issue, as presented in (1), stands at . Meanwhile, the complexity associated with the outer iteration, which is relevant for the convergence of , is represented as .)
To illustrate our contributions, we present a comparison of our mini-batch approach with highly related existing mini-batch methods in the literature. Notably, the studies by [7, 8] introduce SGD techniques for hyperparameter learning in GP regressions. A key distinction of our method lies in its treatment of the minimization over the latent vector , as outlined in (1.2). In contrast, [7, 8] focus on optimizing kernel hyperparameters for given data , without considering this minimization. In alignment with our theoretical findings, the studies [7, 8] similarly identify errors comprising two main elements: an optimization error term and a statistical error term. These are determined by the number of optimization iterations and the mini-batch size. Consequently, with a fixed batch size, it becomes challenging to achieve zero error as the number of optimization steps grows. A key reason that we cannot eliminate the statistical error is the exclusion of a proximal step for as seen in (1). If we were to integrate the proximal step for , it would require maintaining all mini-batch samples and making the iteration’s minimizer depending on the expressions of all previous minimizers . This integration would lead to significantly higher memory demands (see also Remark 3.7). Exploring ways to reduce statistical error while balancing memory efficiency remains an area for future investigation.
Furthermore, [45] develops a stochastic Gauss-Newton algorithm for non-convex compositional optimization. However, their approach is not applicable in our context due to the quadratic nature of (1.2), which is incompatible with their algorithm’s requirement for an expectation-based formulation in finite-dimensional settings. This is evident as the term in Equation (1.2) cannot be framed as an expectation over . Our method employs a mini-batch approach solving the infinite-dimensional minimization problem in (1.1).
Another subtle difference is our methodological approach compared to that of [45],. While they initiate their process by linearizing the objective function and then applying mini-batch updates to this linearized problem, we start by formulating a nonlinear proximal minimization step in each iteration. For solving this, we primarily use the Gauss-Newton method. It’s important to note, however, that the Gauss-Newton method in our framework can be substituted with any other efficient optimization algorithm suitable for the proximal iteration step.
We organize the paper as follows. Section 2 summarizes the GP method in [9] and proposes a mini-batch method to solve PDEs in a general setting. In Section 3, we extend the arguments in the framework of [12, 6, 13] and present a convergence analysis of the min-batch algorithm based on stability analysis and convexity arguments. Numerical experiments appear in Section 4. Further discussions and future work appear in Section 5.
Notations A0.
Given an index set , we denote by the cardinality of . For a real-valued vector , we represent by the Euclidean norm of and by its transpose. Let or be the inner product of vectors and . For a normed vector space , let be the norm of . Let be a Banach space associated with a quadratic norm and let be the dual of . Denote by the duality pairing between and , and by the inner produce of . Suppose that there exists a linear, bijective, symmetric (), and positive ( for ) covariance operator , such that . Let be a set of elements in and let be in the product space . Then, for , we write . Let . Furthermore, for , , let be the matrix with entries . Finally, we represent by a positive real number whose value may change line by line.
2. GP Regressions with Nonlinear Measurements
For ease of presentation, in this section, we introduce an iterative mini-batch method for solving nonlinear PDEs in the general setting of GP regressions with nonlinear measurements. We first summarize the GP method proposed by [9] in Subsection 2.1. Then, in Subsection 2.2, we detail the min-batch method and derive a representer theorem, from which we see that the mini-batch method allocates the burden of computations to each iteration.
2.1. A Revisit of The GP Method
Here, we summarize the general framework of the GP method [9] for solving nonlinear PDEs. Let be an open bounded domain with the boundary for . Let be bounded linear operators for , where . We solve
| (2.1) |
where is the unknown function, and represent nonlinear operators, and , are given data. Throughout this paper, we suppose that (2.1) admits a unique strong solution in a RKHS associated with the covariance operator .
To approximate in , we first take sample points in such that and for . Then, we define
Let be the vector concatenating the functionals for fixed and define
| (2.2) |
Next, we define the data vector by
Furthermore, we define the nonlinear map
Then, given a regularization parameter , the GP method [9] approximates by the minimizer of the following optimization problem
| (2.3) |
The authors in [9] show that as and as the fill distance of samples decreases, a minimizer of (2.3) converges to the unique strong solution of (2.1) (see Sec. 3 of [9]).
2.2. GP Regressions with Nonlinear Measurements
In this subsection, we propose a mini-batch method to solve (2.3). For ease of presentation, we abstract (2.3) into a general framework by considering a GP problem with nonlinear observations. More precisely, let and , where for some and . For an unknown function and each , there exists a function such that we get a noisy observation on . To approximate based on the nonlinear observations, we solve
| (2.4) |
where . Hence, (2.4) resembles (2.3) in such a way that corresponds to the PDE equation satisfied by at the sample point.
As we mentioned before, a typical algorithm using the Cholesky decomposition to compute the inverse of the corresponding covariance matrix suffers the cost of . Here, we introduce an algorithm to solve (2.4) using mini-batches such that the dimensions of matrices to be inverted depend only on the sizes of mini-batches.
First, we reformulate a relaxed version of (2.4). More precisely, we introduce slack variables , a regularization parameter , and consider the following regularized optimal recovery problem
| (2.5) |
where
| (2.6) |
is a convex compact subset of , , and . We further assume that is the product space of a set of convex compact subsets , where .
Remark 2.1.
The restriction of on is not restrictive. Indeed, we can reformulate the unconstrained minimization problem
| (2.7) |
into the formulation of (2.5) by introducing a large enough compact set . More precisely, let be the minimizer of (2.7). We have
which implies that and are bounded if . Hence, if are bounded, (2.7) and (2.5) are equivalent for large . Thus, we mainly focus on (2.5) to take into consideration situations when is constrained.
Next, we introduce a mini-batch version of defined in (2.5). For with , we define
| (2.8) |
In particular, when and for , we denote
Then, we detail the mini-batch method in Algorithm 1. Given and the iteration number , at the step, we update by solving the minimization problem (2.10). Finally, we uniformly sample from and output as an approximation to a solution of (2.5).
Remark 2.2.
In Algorithm 1, we introduce the last step (2.11) for error analysis. In practice, we do not need to solve (2.11) directly since computing (2.11) consumes the same computational cost as calculating (2.4). Instead, if we are interested in the value of at a test point , we can take a mini-batch containing training samples at the neighbor of , choose and as in Algorithm 1, compute , and evaluate at . An alternative way is to include the test points of interest in the training samples and solve (2.5) using the mini-batch method. Then, the latent vector contains sufficient information at .
Remark 2.3.
Algorithm 1 can be readily extended to incorporate a preconditioned variant. Specifically, let be a positive definite matrix associated with the iteration. The update step in Equation (2.10) is modified as follows:
| (2.9) |
where the norm is defined by . This preconditioned approach maintains the theoretical foundations of Algorithm 1, given that the minimum eigenvalue of meets the analogous conditions set by in (2.10) for each iteration . This adaptation offers enhanced flexibility and potential efficiency improvements in various computational scenarios.
| (2.10) |
| (2.11) |
Though the minimization problem in (2.10) is infinite-dimensional, we can reformulate (2.10) into a finite-dimensional minimization problem by using the framework developed in [43]. The following lemma resembles Lemma 3.3 in [29] and establishes properties of a sampling operator, which are crucial for us to derive a representer formula for (2.10). We omit the proof since the arguments are the same as those for Lemma 3.3 of [29]. For simplicity, we denote by the identity map or the identity matrix, whose meaning is easily recognizable from the context.
Lemma 2.4.
Let and . Let be as in (2.4) and let be the subset of indexed by . Denote by the set of sequences with the inner product . Define the sampling operator by . Let be the adjoint of . Then, for each , . Besides, for any , and are bijective. Meanwhile, for any , we have
| (2.12) |
Furthermore,
| (2.13) |
The next lemma provides a representer formula for (2.10). The arguments are similar to those for Theorem 3.4 of [29], which we postpone to Appendix A. We recall that for an index set and a vector , represents the vector consisting of elements of indexed by .
Lemma 2.5.
The above lemma establishes the solvability of (2.10) and implies that in each step of Algorithm 1, the sizes of the matrices to be inverted depend only on the batch size . Thus, the computational cost is reduced to if we use the standard Cholesky decomposition. In numerical implementations of solving PDEs, an alternative way to deal with the PDE constraints in (2.8) at each step is to use the technique of eliminating variables (see Subsection 3.3 of [9] for details), which further reduces the dimensions of unknown variables.
Remark 2.6.
In numerical experiments, we replace the term by a nugget , where is a small real number and is a block diagonal matrix formulated using the method described in Sec. 3.4 of [9]. Using block diagonal matrices lets us choose smaller nuggets and get more accurate results.
3. The Convergence Analysis
In this section, we study the convergence of Algorithm 1. According to (2.15), at the step of Algorithm 1, is expressed as the linear combinations of , where only contains linear operators corresponding to a mini-batch, which is not expressive. Thus, at the final step, we do not expect that approximates a minimizer of (2.5) very well. Instead, we study the accuracy of the sequence in Algorithm 1, which contains the approximated values of the linear operators of the true solution, and we study the convergence of in (2.11), which uses the information of all the linear operators.
We first summarize some basic notions of convex analysis in Subsection 3.1. Then, we introduce the stability of Algorithm 1 in Subsection 3.2. The convergence analysis follows in Subsection 3.3. The arguments are based on the framework of [12] and stability analysis [6]. Similar arguments are used in [13] to analyze the convergence of algorithms for general weakly convex optimization problems.
3.1. Prerequisites
For a non-empty, compact, and convex set for some , the normal cone to at is defined by . Given , a function is -weakly convex if the map is convex and is -strongly convex if is convex. We recall that the proximal map of is given by , and that the Moreau envelop [30] is defined as . Standard results ([30] and [40, Theorem 31.5]) implies that if is -weakly convex and , the envelope is smooth with the gradient given by . Let . Then, the definition of the Moreau envelope implies that . Hence, a point with a small gradient lies in the neighbor of a nearly stationary point .
The following lemma establishes a crucial inequality for the convergence analysis.
Lemma 3.1.
Assume that is -strongly convex in and -weakly convex in . Let and . Then, for any , we have
| (3.1) |
3.2. Stability
In this subsection, we define the notion of stability for (2.10) in a general setting. Let be a set of i.i.d. samples from , where and . Let . We replace with an i.i.d. copy and define . Denote . Let be a random sample from , let be a stochastic model, and let . Given , we define an operator by
| (3.4) |
Given , let . Then, we say that the operator is -stable for some if . The stability measures the accuracy of against the perturbation of one sample. The next lemma shows that if is -stable, the approximation of using a mini-batch is bounded by on expectation. The arguments are similar to those of Lemma 11 of [6] and Theorem A.3 of [13].
Lemma 3.2.
Let be a uniform sample from . For a stochastic model , let be as in (3.4) given . Suppose that is -stable. Let for . Then, we have
| (3.5) |
3.3. Convergence
In this subsection, we study the convergence of Algorithm 1. We first assume that all the linear operators in (2.6) are bounded.
Assumption 1.
Let be the linear operators in (2.6). Assume that there exists a constant such that .
Remark 3.3.
The boundedness imposed on the linear operators is not restrictive if the kernel associated with the space is smooth enough. Indeed, let , where . Then, there exists a constant such that
where we use the reproducing property in the third equality, the Cauchy–Schwarz inequality in the first inequality, and the regularity of the kernel in the final inequality. Similar arguments hold for to be other differential operators.
Furthermore, we impose the following assumption on the regularity of in .
Assumption 2.
Let be functions in (2.6). For any , we assume that is continuous differentiable. Moreover, there exist constants and such that for and
| (3.7) |
With the boundedness of the second derivative of at hand, we are able to show that in (2.6) and in (2.8) are weakly convex. The proof of the following lemma is similar to the argument of Lemma 4.2 of [14]. Recall that is a convex compact set and is the product space of a set of convex compact sets .
Lemma 3.4.
Proof.
For , let and . Using Assumption 2, we obtain
which implies that is -weakly convex. Thus, for , we have
Hence, is also -weakly convex. ∎
Lemma 3.4 directly implies the weakly convexity of defined in (2.8), which is described in the following corollary.
Corollary 3.5.
Next, we use the stability analysis and the weakly convexity property to study the convergence of Algorithm 1. The following lemma establishes the stability for each iteration of Algorithm 1. The proof appears in Appendix A.
Lemma 3.6.
Remark 3.7.
Lemma 3.6 demonstrates the critical distinction between the mini-batch method delineated in this paper and those explored in [15, 12, 13]. In our approach, the regularization parameter is typically small, rendering equivalent to , a fixed parameter that is not dependent on the step size. This aspect is significantly different from the methodologies in [15, 12, 13], where increasing with the number of iterations enables a reduction in statistical error. A notable difference in our method is the omission of a proximal step over in (2.10). Incorporating the proximal step over would necessitate the retention of all mini-batch samples and the dependency of the minimizer at the step on the formulations of preceding minimizers, , thereby substantially elevating memory requirements. Addressing the challenge of reducing statistical error while managing memory efficiency is a subject for future research.
Next, we study the descent property of Algorithm 1. Henceforth, we use the notation to represent the expectation conditioned on all the randomness up to the step in Algorithm 1. The proof appears in Appendix A.
Proposition 3.8.
Before we get to the main theorem, the following corollary implies that in (3.11) is differentiable, which is a direct byproduct of Lemma 2.5.
Corollary 3.9.
Proof.
Next, we have our main theorem about the convergence of Algorithm 1 in the setting of nonlinear measurements.
Theorem 3.10.
Suppose that Assumptions 1 and 2 hold. Let . In Algorithm 1, let for some . In each iteration, let for all , where . Let be the sequence generated by Algorithm 1, let be an index uniformly sampled from , and let be defined as in (3.11) for . Then, there exists a constant depending on such that
| (3.14) |
with , where is defined as in (3.10).
Proof.
Taking , we define as in (3.10). By Corollary 3.9, we get . Thus, using Proposition 3.8, we obtain
| (3.15) |
where is given in (3.9). Summing (3.15) from to , taking the expectation over all the randomness, and dividing the result inequality by , we get
| (3.16) |
Therefore, using the definition of and Jenssen’s inequality, we conclude from (3.16) and (3.9) that (3.14) holds. ∎
Theorem 3.10 shows that tends close to the neighbor of a nearly stationary point on expectation as the number of iterations and the batch size increase. The bound provided in (3.14) demonstrates that the error does not diminish to zero with an increasing number of iterations for a fixed batch size. Conversely, the accuracy improves as the size of mini-batches increases, a fact supported by the subsequent numerical experiments discussed in the following section.






4. Numerical Experiments
In this section, we demonstrate the efficacy of the mini-batch method by solving a nonlinear elliptic equation in Subsection 4.1 and Burgers’ equation in Subsection 4.2. Our implementation uses Python with the JAX package for automatic differentiation. Our experiments are conducted only on CPUs. To better leverage the information between points in each mini-batch, we take mini-batches with nearby samples instead of sampling uniform mini-batches. A mini-batch consists of a uniformly randomly sampled point and its nearest points. We use - tree to support such search, which finds nearest for each point in the whole data set of size in time and space.
4.1. Solving A Nonlinear Elliptic PDE
In this example, we solve a nonlinear elliptic equation by utilizing the preconditioned mini-batch method delineated in Remark 2.3. The chosen preconditioner, , is structured as a diagonal matrix. Its diagonal elements are the diagonal entries of the covariance matrix, specific to each set of mini-batch points. Let and . Given a continuous function , we find solving
| (4.1) |
We prescribe the true solution to be for and compute at the right-hand side of the first equation of (4.1) accordingly. For regression, we choose the Gaussian kernel with the lengthscale parameter . We take samples in the domain with interior points. Meanwhile, we set the nugget (see Remark 2.6). The algorithm stops after iterations. In each time step, we use the technique of eliminating variables at each iteration (see Subsection 3.3 in [9]) and solve the resulting nonlinear minimization problem using the Gauss–Newton method, which terminates once the error between two successive steps is less than or the maximum iteration number is reached.
Figure 1 shows the results of a realization of the mini-batch GP method when the batch size . Figure 1a shows the samples taken in the domain and the contour of the true solution. Figure 1b depicts the reduction in loss for the mini-batch GP method during a single run. The loss is derived from the average of the most recent 10 iterations in the algorithm.. The explicit solution of is given in Figure 1c. Figure 1d plots the approximation of , denoted by . The contour of the point-wise errors of and is given in Figure 1e. Figure 1f presents the contour of errors for the GP method as described in [9], using the whole identical sample sets. It is observed that the mini-batch approach with a batch size of 64 achieves accuracy comparable to that of the GP method using the entire samples.
4.2. Burgers’ Equation
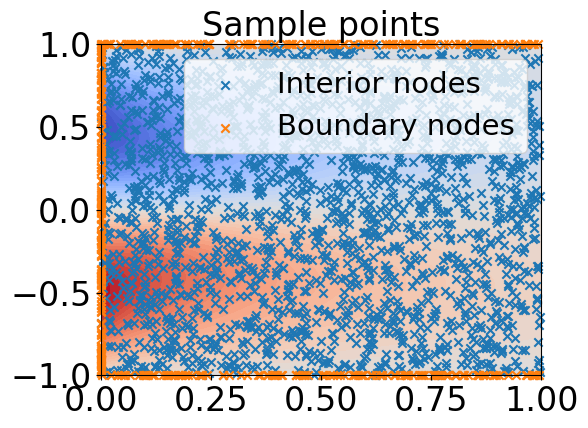




In this subsection, we consider Burgers’ equation
where and is unknown. We take points uniformly in the space-time domain, of which points are in the domain’s interior. As in [9], we choose the anisotropic kernel
| (4.2) |
with . To compute pointwise errors, as in [9], we compute the true solution using the Cole–Hopf transformation and the numerical quadrature. We use the technique of eliminating constraints at each iteration (see Subsection 3.3 in [9]) and use the Gauss–Newton method to solve the resulting nonlinear quadratic minimization problem, which terminates once the error between two successive steps is less than or the maximum iteration number is reached. Throughout the experiments, we use the nugget (see Remark 2.6).




Figure 2 shows the results of a realization of the mini-batch GP method when we fix the batch size . Figure 2a shows the samples in the domain and the contour of the true solution. Figure 2b illustrates the history of the loss generated by the min-batch GP method in one realization. The explicit solution of is plotted in Figure 2c. Figure 2d shows the numerical approximation of , denoted by . The contour of the point-wise errors between and is presented in Figure 2e.
Then, we run the algorithm times for several batch sizes and plot the results in Figure 3. The averaged loss histories are plotted in Figure 3a, from which we see that increasing batch sizes results in faster convergence rates and smaller losses. The contours of averaged point-wise errors are presented in Figures 3b-3d, which imply that a larger batch size achieves better accuracy.
In our observations regarding the solution of Burgers’ equation, it becomes evident that a larger number of samples in a mini-batch is required compared to the nonlinear elliptic example. This disparity could be attributed to the relatively lower regularity of Burgers’ equation, where the uniformly distributed samples may not adequately capture the inherent regularity. Consequently, we defer the exploration of more effective strategies for sampling collocation points and selecting mini-batch points for future investigations.
5. Conclusions
This paper presents a mini-batch framework to solve nonlinear PDEs with GPs. The mini-batch method allocates the computational cost of GPs to each iterative step. We also perform a rigorous convergence analysis of the mini-batch method. For future work, extending our framework to more general GP regression problems like semi-supervised learning and hyperparameter learning would be interesting. In the numerical experiments, we notice that the choice of the positions of mini-batch samples influences the performance. Hence, a potential future work is investigating different sampling techniques to take mini-batch sample points.
Acknowledgement A0.
The authors wish to thank Professor Andrew Stuart for his insightful discussions on the convergence of the stochastic gradient descent method.
A Proofs of Results
Proof of Lemma 2.5.
Let . Define the sampling operator as in Lemma (2.4). Then, we have
| (A.1) |
Next, we first fix and let . Then,
which yields
| (A.2) |
Thus, using (A) and (A.2), we have
| (A.3) |
where the last equality results from (2.13). Therefore, (A.2), (2.12), and (A.3) imply that the minimizer in (2.14) satisfies (2.15) and minimizes (2.16). ∎
Proof of Lemma 3.6.
Let and be as in the beginning of this subsection. For , let
| (A.4) |
For , by the optimality condition of (A.4), there exists a constant depending on , , the diameter of , and the constant in Assumption 2, such that
| (A.5) |
Thus, using (A.5) and Assumption 1, there exists a constant depending on , , , , and the constant in Assumption 1, such that . Let , where is large enough such that contains in (A.4) for any . Next, we show that for any , is Lipschitz in . Let . Then, there exists a constant depending on , and , such that
| (A.6) |
where (A.6) follows by the definition of and Assumptions 1 and 2.
By Jensen’s inequality, we obtain
| (A.7) |
where (A) follows by (A.6). Using (A.4), Corollary 3.5, and Lemma 3.1, we get
| (A.8) | ||||
| (A.9) |
Adding (A.8) and (A.9) together and using (A.6), we obtain
which together with the fact that , yield
| (A.10) |
Hence, (A) and (A.10) imply that is -stable with given in (3.9). ∎
Proof of Proposition 3.8.
Let be a mini-batch of indices in the step of Algorithm 1. According to Corollary 3.5, is -weakly convex in and strongly convex in . Then, by Lemma 3.1 and (2.10), for any and , we have
| (A.11) |
| (A.12) |
where is given in (3.9) with . For , let be given in (3.10). Then, we have
| (A.13) |
Plugging into (A.11), taking the expectation, using (A.12) and (A.13), and using Lemma 3.6, we obtain
| (A.14) |
For , using the definition of and (A.14), we have
which concludes (3.8) after rearrangement. ∎
References
- [1] H. Asi and J. C. Duchi, The importance of better models in stochastic optimization, Proceedings of the National Academy of Sciences, 116 (2019), pp. 22924–22930.
- [2] H. Asi and J. C. Duchi, Stochastic (approximate) proximal point methods: Convergence, optimality, and adaptivity, SIAM Journal on Optimization, 29 (2019), pp. 2257–2290.
- [3] P. Batlle, Y. Chen, B. Hosseini, H. Owhadi, and A. M. Stuart, Error analysis of kernel/GP methods for nonlinear and parametric PDEs, arXiv preprint arXiv:2305.04962, (2023).
- [4] D. P. Bertsekas, Incremental proximal methods for large scale convex optimization, Mathematical programming, 129 (2011), pp. 163–195.
- [5] L. Bottou and O. Bousquet, The tradeoffs of large scale learning, Advances in neural information processing systems, 20 (2007).
- [6] O. Bousquet and A. Elisseeff, Stability and generalization, The Journal of Machine Learning Research, 2 (2002), pp. 499–526.
- [7] H. Chen, L. Zheng, R. Al Kontar, and G. Raskutti, Stochastic gradient descent in correlated settings: A study on Gaussian processes, Advances in neural information processing systems, 33 (2020), pp. 2722–2733.
- [8] H. Chen, L. Zheng, R. Al Kontar, and G. Raskutti, Gaussian process parameter estimation using mini-batch stochastic gradient descent: convergence guarantees and empirical benefits, The Journal of Machine Learning Research, 23 (2022), pp. 10298–10356.
- [9] Y. Chen, B. Hosseini, H. Owhadi, and A. M. Stuart, Solving and learning nonlinear PDEs with Gaussian processes, Journal of Computational Physics, (2021).
- [10] Y. Chen, H. Owhadi, and F. Schäfer, Sparse Cholesky factorization for solving nonlinear PDEs via Gaussian processes, arXiv preprint arXiv:2304.01294, (2023).
- [11] A. C. Damianou, M. K. Titsias, and N. D. Lawrence, Variational inference for latent variables and uncertain inputs in Gaussian processes, (2016).
- [12] D. Davis and D. Drusvyatskiy, Stochastic model-based minimization of weakly convex functions, SIAM Journal on Optimization, 29 (2019), pp. 207–239.
- [13] Q. Deng and W. Gao, Minibatch and momentum model-based methods for stochastic weakly convex optimization, Advances in Neural Information Processing Systems, 34 (2021), pp. 23115–23127.
- [14] D. Drusvyatskiy and C. Paquette, Efficiency of minimizing compositions of convex functions and smooth maps, Mathematical Programming, 178 (2019), pp. 503–558.
- [15] J. C. Duchi and F. Ruan, Stochastic methods for composite and weakly convex optimization problems, SIAM Journal on Optimization, 28 (2018), pp. 3229–3259.
- [16] J. Gardner, G. Pleiss, K. Q. Weinberger, D. Bindel, and A. G. Wilson, GPytorch: Blackbox matrix-matrix Gaussian process inference with GPU acceleration, Advances in neural information processing systems, 31 (2018).
- [17] J. Hensman, N. Durrande, and A. Solin, Variational Fourier features for Gaussian processes., J. Mach. Learn. Res., 18 (2017), pp. 5537–5588.
- [18] J. Hensman, N. Fusi, and N. D. Lawrence, Gaussian processes for big data, in Proceedings of the Twenty-Ninth Conference on Uncertainty in Artificial Intelligence, UAI’13, Arlington, Virginia, United States, 2013, AUAI Press, pp. 282–290, http://dl.acm.org/citation.cfm?id=3023638.3023667.
- [19] T. J. Hughes, The finite element method: linear static and dynamic finite element analysis, Courier Corporation, 2012.
- [20] G. E. Karniadakis, I. G. Kevrekidis, L. Lu, P. Perdikaris, S. Wang, and L. Yang, Physics-informed machine learning, Nature Reviews Physics, 3 (2021), pp. 422–440.
- [21] N. Krämer, J. Schmidt, and P. Hennig, Probabilistic numerical method of lines for time-dependent partial differential equations, arXiv preprint arXiv:2110.11847, (2021).
- [22] B. Kulis and P. L. Bartlett, Implicit online learning, in Proceedings of the 27th International Conference on Machine Learning (ICML-10), 2010, pp. 575–582.
- [23] M. Lázaro-Gredilla and A. Figueiras-Vidal, Inter-domain Gaussian processes for sparse inference using inducing features, Advances in Neural Information Processing Systems, 22 (2009).
- [24] M. Lázaro-Gredilla, J. Quinonero-Candela, C. E. Rasmussen, and A. R. Figueiras-Vidal, Sparse spectrum Gaussian process regression, The Journal of Machine Learning Research, 11 (2010), pp. 1865–1881.
- [25] L. Li, K. Jamieson, G. DeSalvo, A. Rostamizadeh, and A. Talwalkar, Hyperband: A novel bandit-based approach to hyperparameter optimization, The Journal of Machine Learning Research, 18 (2017), pp. 6765–6816.
- [26] H. Liu, Y. S. Ong, X. Shen, and J. Cai, When Gaussian process meets big data: A review of scalable GPs, IEEE transactions on neural networks and learning systems, 31 (2020), pp. 4405–4423.
- [27] L. Lu, P. Jin, G. Pang, Z. Zhang, and G. E. Karniadakis, Learning nonlinear operators via deeponet based on the universal approximation theorem of operators, Nature Machine Intelligence, 3 (2021), pp. 218–229.
- [28] Z. Mao, A. D. Jagtap, and G. E. Karniadakis, Physics-informed neural networks for high-speed flows, Computer Methods in Applied Mechanics and Engineering, 360 (2020), p. 112789.
- [29] R. Meng and X. Yang, Sparse Gaussian processes for solving nonlinear PDEs, Journal of Computational Physics, 490 (2023), p. 112340.
- [30] J.-J. Moreau, Proximité et dualité dans un espace hilbertien, Bulletin de la Société mathématique de France, 93 (1965), pp. 273–299.
- [31] C. Mou, X. Yang, and C. Zhou, Numerical methods for mean field games based on Gaussian processes and Fourier features, Journal of Computational Physics, (2022).
- [32] A. Nemirovski, A. Juditsky, G. Lan, and A. Shapiro, Robust stochastic approximation approach to stochastic programming, SIAM Journal on optimization, 19 (2009), pp. 1574–1609.
- [33] D. T. Nguyen, M. Filippone, and P. Michiardi, Exact Gaussian process regression with distributed computations, in Proceedings of the 34th ACM/SIGAPP symposium on applied computing, 2019, pp. 1286–1295.
- [34] H. Owhadi and C. Scovel, Operator-Adapted Wavelets, Fast Solvers, and Numerical Homogenization: From a Game Theoretic Approach to Numerical Approximation and Algorithm Design, vol. 35, Cambridge University Press, 2019.
- [35] A. Quarteroni and A. Valli, Numerical approximation of partial differential equations, vol. 23, Springer Science and Business Media, 2008.
- [36] J. Quinonero-Candela and C. E. Rasmussen, A unifying view of sparse approximate Gaussian process regression, The Journal of Machine Learning Research, 6 (2005), pp. 1939–1959.
- [37] A. Rahimi and B. Recht, Random features for large-scale kernel machines., in NIPS, vol. 3, Citeseer, 2007, p. 5.
- [38] M. Raissi, P. Perdikaris, and G. E. Karniadakis, Numerical Gaussian processes for time-dependent and nonlinear partial differential equations, SIAM Journal on Scientific Computing, 40 (2018), pp. A172–A198.
- [39] M. Raissi, P. Perdikaris, and G. E. Karniadakis, Physics-informed neural networks: A deep learning framework for solving forward and inverse problems involving nonlinear partial differential equations, Journal of Computational physics, 378 (2019), pp. 686–707.
- [40] R. Rockafellar, Convex analysis, vol. 18, Princeton university press, 1970.
- [41] F. Schäfer, M. Katzfuss, and H. Owhadi, Sparse Cholesky factorization by Kullback–Leibler minimization, SIAM Journal on scientific computing, 43 (2021), pp. A2019–A2046.
- [42] S. Shai, Y. Singer, and N. Srebro, Pegasos: Primal estimated sub-gradient solver for SVM, in Proceedings of the 24th international conference on Machine learning, 2007, pp. 807–814.
- [43] S. Smale and D. Zhou, Shannon sampling II: Connections to learning theory, Applied and Computational Harmonic Analysis, 19 (2005), pp. 285–302.
- [44] J. W. Thomas, Numerical partial differential equations: finite difference methods, vol. 22, Springer Science and Business Media, 2013.
- [45] Q. Tran-Dinh, N. Pham, and L. Nguyen, Stochastic gauss-newton algorithms for nonconvex compositional optimization, in International Conference on Machine Learning, PMLR, 2020, pp. 9572–9582.
- [46] J. Wang, J. Cockayne, O. Chkrebtii, T. J. Sullivan, and C. J. Oates, Bayesian numerical methods for nonlinear partial differential equations, Statistics and Computing, 31 (2021), pp. 1–20.
- [47] K. Wang, G. Pleiss, J. Gardner, S. Tyree, K. Q. Weinberger, and A. G. Wilson, Exact Gaussian processes on a million data points, Advances in neural information processing systems, 32 (2019).
- [48] A. Wilson and H. Nickisch, Kernel interpolation for scalable structured Gaussian processes (KISS-GP), in International conference on machine learning, PMLR, 2015, pp. 1775–1784.
- [49] F. X. Yu, A. T. Suresh, K. M. Choromanski, D. N. Holtmann-Rice, and S. Kumar, Orthogonal random features, Advances in neural information processing systems, 29 (2016), pp. 1975–1983.
- [50] M. Zinkevich, Online convex programming and generalized infinitesimal gradient ascent, in Proceedings of the 20th international conference on machine learning (ICML-03), 2003, pp. 928–936.