A hybrid convolutional neural network/active contour approach to segmenting dead trees in aerial imagery
Abstract
The stability and ability of an ecosystem to withstand climate change is directly linked to its biodiversity. Dead trees are a key indicator of overall forest health, housing one-third of forest ecosystem biodiversity, and constitute of the global carbon stocks. They are decomposed by several natural factors, e.g. climate, insects and fungi. Accurate detection and modeling of dead wood mass is paramount to understanding forest ecology, the carbon cycle and decomposers. We present a novel method to construct precise shape contours of dead trees from aerial photographs by combining established convolutional neural networks with a novel active contour model in an energy minimization framework. Our approach yields superior performance accuracy over state-of-the-art in terms of precision, recall, and intersection over union of detected dead trees. This improved performance is essential to meet emerging challenges caused by climate change (and other man-made perturbations to the systems), particularly to monitor and estimate carbon stock decay rates, monitor forest health and biodiversity, and the overall effects of dead wood on and from climate change.
1 Introduction
With the increasing global interest in understanding and mitigating climate change, researchers find themselves presented with new problems. One such problem is understanding the role and behavior of dead trees in these processes, as they are a key indicator of forest health. Forests are a core component in the global carbon cycle and are the most efficient ecosystem on the planet for scrubbing CO2 and returning oxygen to the atmosphere, sequestering as much CO2 as all of the oceans. Carbon stocks and fluxes in dead wood – fallen and standing dead trees, branches, and other woody tissues – are a critical component of forest carbon dynamics [1], constituting of the global forest carbon stocks [2]. Furthermore, dead wood houses one-third of all forest biodiversity, which is of crucial importance as an ecosystem’s bioversity is directly linked to its stability and the ability to withstand climate change. Dead trees are decomposed by several natural factors (including climate, fungi and insects), however, the influence of these decomposers as well as the impact of environmental change upon them remains poorly understood. While initial studies of both insects [3] and fungi [4, 5] have been performed, further studies are still needed to gain a more holistic understanding. In particular, there is an increasing need for both larger scale and longitudinal studies of the impact of dead trees on the ecology of forests, and their interaction with the carbon cycle and decomposers (see e.g. [1] and citations within). These efforts are hindered by a lack of data and tools for processing the data, particularly from aerial photography, which offers a good trade-off between high spatial resolution and cost efficiency, making it ideal for localized studies. In order to address this need we propose the use of Machine Learning (ML) algorithms to identify the location and shape of dead trees. Namely, using Computer Vision (CV) ML techniques applied to aerial photos of a forest at multiple time steps a temporal change in tree crowns can be made, providing estimates in decay rates.
The motivation of this work is to develop a method to accurately model and estimate dead wood mass. There are however other applications ranging e.g. from tracking the health of a forest by identifying dead and dying trees from invasive insects and disease, to tracking desertification and reforestation after harvesting or wildfires, to the development of algal blooms in the ocean. The precise fallen tree maps could be further used as a basis for plant and animal habitat modeling, studies on carbon sequestration as well as soil quality in forest ecosystems.
The method we propose is a hybrid of two convolutional neural networks with a novel active contour model for precise object contour segmentation. We use infrared aerial imagery to identify dead vegetation, a widely used technique due to the difference in reflectance caused by differences in chlorophyll in the near-infrared spectral band. Due to recent improvements in this technology, in specific the increase in resolution, the current existent, non-Machine Learning, methods are unable to provide the highest satisfactory performance. These discrepancies are then only exaggerated when the amount of available data is drastically increased by the use of unmanned aerial vehicles (UAVs) to collect data more often for the same forest. The details of the method are as follows: We use leading convolutional network approaches U-Net for instance segmentation to compute class probability masks of the dead trees and a Mask R-CNN (Mask Regional-CNN) to segment the image into components, in particular separating trees from each other. The Mask R-CNN can successfully identify the number and precise position of the trees. To further improve the contours of the dead trees we then apply a contour refinement step based on a generalized classical computer vision technique by using simultaneously evolving contours based upon energy functions.
The goal of the present work is to design a cutting edge Machine Learning algorithm for identifying dead trees in a forest, and then determining the shape and location of the dead tree’s crown. With this information, crucial aspects of carbon decay can be more accurately estimated and predicted. Experimental results yield superior performance over conventional instance segmentation methods, reducing the cost of large scale studies allowing for improved understanding of forest health, and how that is impacted through time by factors such as insects, natural disturbances, and especially climate change. The paper is organized as follows: Sec. 2 introduces the proposed hybrid method, Sec. 3 presents experimental results, and finally Sec. 4 provides a summary of the work and outlook.
2 A hybrid approach to contour modeling of dead trees in aerial images
We first describe the convolutional neural networks implemented for instance segmentation and object localization. Then we introduce our main technical contribution which harnesses the advantages of these networks to construct our method. Specifically, the U-Net gives us the probabilities of which pixels belong to which dead trees (classes) and the Mask R-CNN provides solid estimates of the locations of each tree (centroids). We combine these results in a novel energy minimization framework for high resolution contour modeling. Figure 1 provides a simplified overview of the entire process.

U-Net. The U-Net [6] is a fully convolutional neural network architecture which constitutes a milestone in the task of image dense semantic segmentation. This network is particularly well suited for our problem because it preserves the object contours well, an imperative aspect for retaining fine details of the tree crowns.
Mask R-CNN. Mask R-CNN [7], or Region Based Convolutional Neural Networks, is a state of the art neural network architecture for the task of generic image instance segmentation, i.e. obtaining separate pixel masks for all object instances present within the input image. Mask R-CNN has two stages: (i) a region proposal network, which selects promising image regions that are likely to contain object instances, and (ii) a fine-grained detection component which examines the candidate regions and predicts the object class label, bounding box, and the instance’s pixel mask.
Active contour segmentation with energy minimization. We formalize the above setting with our contour model as follows. Let be the object centroids identified by the M-RCNN, and let denote the dead tree class posterior probability image obtained from the U-Net. Furthermore, let be the image plane, a vector-valued image, and an evolving contour in the image . The one-shape segmentation in the active contour model (ACM) w.r.t. shape and appearance priors consists in finding a contour which ‘optimally’ partitions into disjoint interior and exterior regions such that the probability induced by is minimized [8]:
| (1) |
Furthermore, the contour is parameterized by a vector of shape coefficients and a offset vector . A shape generator is given, which instantiates the contour in standard position and translates the center to . The image term can be interpreted as the pixel-wise cross entropy between the target class posterior probability image and the indicator function of the contour’s interior (see [9] for details). In our setting, we consider an arbitrary number of simultaneously evolving contours , each having its own shape coefficients and offset vector. The image energy term is now defined as the cross entropy between the set-theoretic union of all generated contours and the posterior probability image. Moreover, we introduce a new term into the energy, which penalizes the total pairwise overlap between evolving model shapes, to make sure they cover different regions of the input image. We approximate the overlap between as the product . The final energy formulation can be written as:
| (2) |
In the above expression, the union operation can be implemented by taking the pixel-wise maximum over all generated shapes . However, since the max function is not differentiable, we apply a smooth approximation , where is a positive constant. The coefficients control the balance of terms within the energy function. We utilize the eigenshape model [10, 11] in the role of , whereas the shape probability follows the kernel density estimator model proposed by Cremers et al. [12]. In practice, the optimization requires good initial object positions and the object count . We utilize the centroids obtained from Mask RCNN in this role. The evolving shape positions are constrained to lie within pixels of .
3 Numerical Experiments
Data. We use high resolution aerial images acquired by a flight campaign from the Bavarian Forest National Park in Germany with centimeter ground pixel resolution (see Appendix B.1 for details). We manually marked outlines of dead trees within the color infrared images of a selected area in the National Park (Fig. 5(a)) for training all components of the segmentation pipeline: the U-Net, the Mask R-CNN and the active contour model. We employed a semi-automatic strategy for acquiring dead tree crown polygon testing data. We applied the trained U-Net to a new, previously unseen region of the National Park, and obtained the dead tree crown per-pixel probability map. We subsequently manually partitioned a number of connected components into individual tree crowns by applying split polylines to cut parts off the main polygon (Fig. 5(c)), for a total of artificial tree crown polygons. These polygons were used to validate our approach and compare against the pure Mask R-CNN baseline.
Training the models. For the U-Net, we followed the original architecture proposed in [6], and trained the network for epochs on a total of patches of size pixels. We trained the Mask R-CNN on patches of size until convergence of the validation loss curve ( epochs). (Implementation details can be found in Appendix A.1). The eigenshape model was learned from the training contours for the two CNNs, using top eigenmodes of variation and including rotated and flipped copies of the original polygons.
Contour retrieval performance. We ran several experiments comparing the quality of the extracted dead tree crown masks between the baseline method of Mask R-CNN and the active contour model based refinement. To this end, the aforementioned dead tree crowns were distributed into images of dimensions (same as training patches). We executed the pipeline from Fig.1 on the test images until convergence, yielding refined contours. To solve the (box-)constrained continuous energy minimization problem from Eq. (2), we used the L-BFGS method. To assess the quality of both sets of masks, we used the following metrics: (i) mean centroid distance between reference and detected tree crown masks mean, (ii) Intersection over Union (IoU) [13] of detected vs. reference polygons, and (iii) precision and recall at IoU . The results are visualized in Fig.2. The true centroids of the dead tree crowns can be approximated by the centroids found by Mask R-CNN very well (average deviation of pixels) and thus serve as good seed points for the ACM contour model. The ACM refinement further improved this value by ca. pixel to pixels, as shown in Fig.2. On average, the IoU improved by ca. percentage points (pp) after refinement, leading to an increase in precision and recall by, respectively and pp. Moreover, we observe that as the number of dead trees present within the image increases, the detection recall for the ACM refinement does not drop as quickly as for the baseline method. There were a total of vs. dead trees reported respectively by Mask R-CNN and ACM methods. Sample dead tree crown contours from our ACM approach and Mask R-CNN are shown in Fig.3.


4 Conclusions and Discussion
Dead trees are a key indicator of overall forest health, biodiversity and are a crucial component of forest carbon dynamics that are heavily influenced by climate change, insects, and fungi. In order to aid in understanding e.g. dead wood decomposition, this work proposed a hybrid of two convolutional neural networks (U-Net and Mask R-CNN) with a novel active contour model (ACM) for precise dead tree contour segmentation.
Our numerical experiments comparing our (ACM) approach to Mask R-CNN as a baseline show that although the latter yields good estimates of the number and location of dead trees in an image, the alignment of the detected contour with the true dead crown is poor. On the other hand, applying the ACM based contour refinement can significantly improve this alignment (by pp on average), as measured by the overlap (IoU). Furthermore, these experiments show that ACM is more robust in the presence of more difficult scenarios as measured by the number of dead trees present in the image.
Future work includes incoorporating contour shape priors that can capture even more fine details of the tree crowns (e.g. generated by GANs) than can the current eigenshape prior. Another focus will be to use the proposed method to improve estimates of dead wood decay rates by means of, e.g. temporal change detection, with which to ideally form models of decay dynamics dependent on different factors, e.g. geographical location and tree species. These additions will further help monitor and understand forest ecosystem health and biodiversity, and the role of dead wood and its impacts on and from our rapidly changing climate.
References
- [1] A. R. Martin, G. M. Domke, M. Doraisami, and S. C. Thomas, “Carbon fractions in the world’s dead wood,” Nature Communications, vol. 12, 2021.
- [2] Y. Pan, R. A. Birdsey, J. Fang, R. Houghton, P. E. Kauppi, W. A. Kurz, O. L. Phillips, A. Shvidenko, S. L. Lewis, J. G. Canadell, P. Ciais, R. B. Jackson, S. W. Pacala, A. D. McGuire, S. Piao, A. Rautiainen, S. Sitch, and D. Hayes, “A large and persistent carbon sink in the world’s forests,” Science, vol. 333, no. 6045, pp. 988–993, 2011.
- [3] S. Seibold, W. Rammer, T. Hothorn, R. Seidl, M. D. Ulyshen, J. Lorz, M. W. Cadotte, D. B. Lindenmayer, Y. P. Adhikari, R. Aragón, S. Bae, P. Baldrian, H. Barimani Varandi, J. Barlow, C. Bässler, J. Beauchêne, E. Berenguer, R. S. Bergamin, T. Birkemoe, G. Boros, R. Brandl, H. Brustel, P. J. Burton, Y. T. Cakpo-Tossou, J. Castro, E. Cateau, T. P. Cobb, N. Farwig, R. D. Fernández, J. Firn, K. S. Gan, G. González, M. M. Gossner, J. C. Habel, C. Hébert, C. Heibl, O. Heikkala, A. Hemp, C. Hemp, J. Hjältén, S. Hotes, J. Kouki, T. Lachat, J. Liu, Y. Liu, Y.-H. Luo, D. M. Macandog, P. E. Martina, S. A. Mukul, B. Nachin, K. Nisbet, J. O’Halloran, A. Oxbrough, J. N. Pandey, T. Pavlíček, S. M. Pawson, J. S. Rakotondranary, J.-B. Ramanamanjato, L. Rossi, J. Schmidl, M. Schulze, S. Seaton, M. J. Stone, N. E. Stork, B. Suran, A. Sverdrup-Thygeson, S. Thorn, G. Thyagarajan, T. J. Wardlaw, W. W. Weisser, S. Yoon, N. Zhang, and J. Müller, “The contribution of insects to global forest deadwood decomposition,” Nature, vol. 597, pp. 77–81, Sep 2021.
- [4] M. A. Bradford, R. J. Warren II, P. Baldrian, T. W. Crowther, D. S. Maynard, E. E. Oldfield, W. R. Wieder, S. A. Wood, and J. R. King, “Climate fails to predict wood decomposition at regional scales,” Nature Climate Change, vol. 4, pp. 625–630, Jul 2014.
- [5] N. Lustenhouwer, D. S. Maynard, M. A. Bradford, D. L. Lindner, B. Oberle, A. E. Zanne, and T. W. Crowther, “A trait-based understanding of wood decomposition by fungi,” Proceedings of the National Academy of Sciences, vol. 117, no. 21, pp. 11551–11558, 2020.
- [6] O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” CoRR, vol. abs/1505.04597, 2015.
- [7] K. He, G. Gkioxari, P. Dollár, and R. Girshick, “Mask r-cnn,” in 2017 IEEE International Conference on Computer Vision (ICCV), pp. 2980–2988, 2017.
- [8] D. Cremers, M. Rousson, and R. Deriche, “A review of statistical approaches to level set segmentation: Integrating color, texture, motion and shape,” Int. J. Comput. Vision, vol. 72, no. 2, pp. 195–215, 2007.
- [9] P. Polewski, J. Shelton, W. Yao, and M. Heurich, “Segmentation of single standing dead trees in high-resolution aerial imagery with generative adversarial network-based shape priors,” The International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, vol. XLIII-B2-2020, pp. 717–723, 2020.
- [10] M. Leventon, W. Grimson, and O. Faugeras, “Statistical shape influence in geodesic active contours,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, vol. 1, pp. 316–323, 2000.
- [11] H.-F. Tsai, J. Gajda, T. F. Sloan, A. Rares, and A. Q. Shen, “Usiigaci: Instance-aware cell tracking in stain-free phase contrast microscopy enabled by machine learning,” SoftwareX, vol. 9, pp. 230–237, 2019.
- [12] D. Cremers and M. Rousson, “Efficient kernel density estimation of shape and intensity priors for level set segmentation,” in Deformable Models, Topics in Biomedical Engineering. International Book Series, pp. 447–460, Springer, New York, 2007.
- [13] P. Jaccard, “Étude comparative de la distribution florale dans une portion des alpes et des jura,” Bulletin del la Société Vaudoise des Sciences Naturelles, vol. 37, pp. 547–579, 1901.
- [14] J. Akeret, C. Chang, A. Lucchi, and A. Refregier, “Radio frequency interference mitigation using deep convolutional neural networks,” Astronomy and Computing, vol. 18, pp. 35–39, 2017.
- [15] W. Abdulla, “Mask R-CNN for object detection and instance segmentation on keras and tensorflow.” https://github.com/matterport/Mask_RCNN, 2017.
Appendix A Appendix: Experiments
A.1 Implementation of U-Net and Mask R-CNN
The tensorflow implementation of the U-Net in [14] was adapted to support masking out irrelevant parts of the images in the training phase. We used the original architecture proposed by [6], and trained the network for epochs on a total of patches of size pixels.
We used the implementation of Mask R-CNN in [15] publicly available on Github. Image augmentation was applied in the form of horizontal and vertical flipping as well as rotation by , and degrees. The optimization on patches of size was conducted until convergence of the validation loss curve ( epochs). Before training the Mask R-CNN, all images were inspected for dead tree crowns which were not labeled. Such tree crowns were overwritten with a neutral color within the image so that detection metrics may be reliably computed (all dead trees detectable within the image are annotated with ground-truth labels).
The eigenshape model was learned from the training contours for the two CNNs, using top eigenmodes of variation and including rotated and flipped copies of the original polygons. The tree crown masks were aligned according to their centroid wihin a 92x92 pixel frame, corresponding to the largest object we wish to detect (crown diameter of 9.2).
A.2 Further refined contour examples



Appendix B Appendix: Data
B.1 Data acquisition
Color infrared images of the Bavarian Forest National Park, situated in South-Eastern Germany ( N, E), were acquired in the leaf-on state during a flight campaign carried out in June 2017 using a DMC III high resolution digital aerial camera.The mean above-ground flight height was ca. 2300 m, resulting in a pixel resolution of 10 cm on the ground. The images contain 3 spectral bands: near infrared, red and green.
B.2 Testing and training data
As mentioned in the Experiments section 2, high resolution aerial data were acquired by a flight campaign from the Bavarian Forest National Park with centimeter ground pixel resolution (see Appendix B.1 for details). We manually marked 201 outlines of dead trees within the color infrared images of a selected area in the National Park (see Fig. 5a). These manually marked polygons were utilized for the purpose of training all components of the segmentation pipeline: the U-Net, the Mask R-CNN and the active contour model. For training the U-Net, we prepared patches of size 200x200 containing the input color infrared image and a pixel mask representing the labeled polygon regions. Also, we constrained the negative class labels to at most 5 pixels away from labeled dead tree polygons, to account for the fact that not all dead tree crowns in the processed images were labeled (Fig. 5b). For training the Mask R-CNN, we utilized 70 patches of size 256x256 with marked individual instances as input (see Fig. 6). Finally, the binary masks of individual marked tree crown polygons were used as a basis for learning the active contour model.



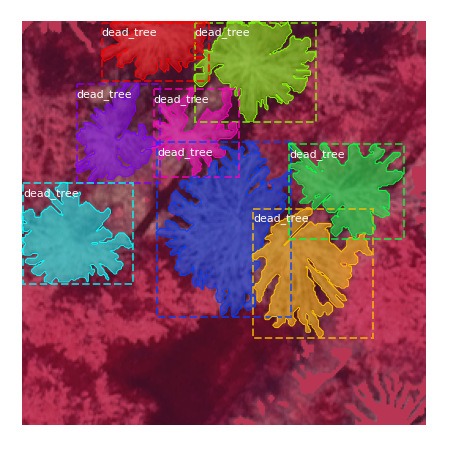
We employed a different, semi automatic strategy for acquiring dead tree crown polygon testing data. We applied the trained U-Net to a new, previously unseen region of the National Park, and obtained the dead tree crown per-pixel probability map. Connected component segmentation was then applied on pixels of the image classified as dead trees. As the test area contained many overlapping and adjacent dead trees, the connected components obtained from this step usually did not represent only single trees, but rather collections of several dead tree crowns. We subsequently manually partitioned a number of connected components into individual tree crowns by applying split polylines to successively cut parts off the main polygon (Fig. 5c). We found this approach to be less time consuming than manually drawing the entire polygons. We obtained a total of 750 artificial tree crown polygons this way. They were utilized for validating our approach and for comparison against the pure Mask R-CNN baseline.